Abstracts
Bovine coronavirus (BCoV) may cause acute diarrhea in newborn calves, leading to significant economic losses for cattle farmers. There are several diagnostic techniques used to detect BCoV in calf fecal samples, but virus isolation still has advantages for antigenic and genomic characterization. This study describes the isolation in HRT-18 cells and molecular characterization of Brazilian BCoV wild-type strains. Three fecal samples from diarrheic 30 day-old calves were inoculated in HRT-18 cell monolayers, which were then evaluated for HA titers and tested using semi-nested PCR followed by RFLP and sequencing. Two samples were successfully isolated and presented HA titers of 16 and 32 units per 25 mL. The results were confirmed using semi-nested PCR and RFLP. Molecular analyses identified a cell culture-adapted strain and a wild-type strain that were genetically similar (99%) to each other, but more distinct than BCoV strains circulating in other countries, even in the conserved N gene.
cattle; diarrhea; bovine coronavirus; cell culture; molecular analyses
O coronavírus bovino (BCoV) pode causar diarreia aguda em bezerros recém-nascidos, ocasionando consideráveis perdas econômicas para a pecuária bovina. Várias técnicas de diagnóstico podem ser empregadas na detecção do BCoV a partir de amostras fecais de bezerros. Porém, o isolamento do BCoV em cultivo celular apresenta a vantagem de possibilitar a caracterização antigênica e molecular da estirpe viral. O presente estudo descreve o isolamento em células HRT-18, e a caracterização molecular de estirpes brasileiras do BCoV. Três amostras de fezes diarreicas de bezerros com 30 dias de idade foram inoculadas em culturas de células HRT-18. Os isolados foram avaliados por hemaglutinação (HA) e por uma semi-nested PCR seguida de RFLP e sequenciamento. Duas amostras foram isoladas e a confirmação foi verificada na semi-nested PCR e também RFLP. Na HA os títulos foram de 16 e 32 unidades por 25 mL. Análises moleculares identificam a estirpe adaptada em cultura celular e uma estirpe selvagem, como estirpes de BCoV semelhantes (99%) entre si, mas distintas das circulantes em outros países, mesmo em um gene de uma proteína conservada (gene N).
HUMAN AND ANIMAL HEALTH
Isolation in HRT-18 cells and molecular analysis of a BCoV strain from diarrheic feces of naturally infected calves
Danilo Tancler Stipp; Aline Fernandes Barry; Alice Fernandes Alfieri; Lívia Bodnar; Amauri Alcindo Alfieri * * Author for correspondence: alfieri@uel.br
Laboratório de Virologia Animal; Departamento de Medicina Veterinária Preventiva; Universidade Estadual de Londrina; C. P.: 6001; 86051-990; Londrina - PR - Brasil
ABSTRACT
Bovine coronavirus (BCoV) may cause acute diarrhea in newborn calves, leading to significant economic losses for cattle farmers. There are several diagnostic techniques used to detect BCoV in calf fecal samples, but virus isolation still has advantages for antigenic and genomic characterization. This study describes the isolation in HRT-18 cells and molecular characterization of Brazilian BCoV wild-type strains. Three fecal samples from diarrheic 30 day-old calves were inoculated in HRT-18 cell monolayers, which were then evaluated for HA titers and tested using semi-nested PCR followed by RFLP and sequencing. Two samples were successfully isolated and presented HA titers of 16 and 32 units per 25 mL. The results were confirmed using semi-nested PCR and RFLP. Molecular analyses identified a cell culture-adapted strain and a wild-type strain that were genetically similar (99%) to each other, but more distinct than BCoV strains circulating in other countries, even in the conserved N gene.
Key words: cattle, diarrhea, bovine coronavirus, cell culture, molecular analyses
RESUMO
O coronavírus bovino (BCoV) pode causar diarreia aguda em bezerros recém-nascidos, ocasionando consideráveis perdas econômicas para a pecuária bovina. Várias técnicas de diagnóstico podem ser empregadas na detecção do BCoV a partir de amostras fecais de bezerros. Porém, o isolamento do BCoV em cultivo celular apresenta a vantagem de possibilitar a caracterização antigênica e molecular da estirpe viral. O presente estudo descreve o isolamento em células HRT-18, e a caracterização molecular de estirpes brasileiras do BCoV. Três amostras de fezes diarreicas de bezerros com 30 dias de idade foram inoculadas em culturas de células HRT-18. Os isolados foram avaliados por hemaglutinação (HA) e por uma semi-nested PCR seguida de RFLP e sequenciamento. Duas amostras foram isoladas e a confirmação foi verificada na semi-nested PCR e também RFLP. Na HA os títulos foram de 16 e 32 unidades por 25 mL. Análises moleculares identificam a estirpe adaptada em cultura celular e uma estirpe selvagem, como estirpes de BCoV semelhantes (99%) entre si, mas distintas das circulantes em outros países, mesmo em um gene de uma proteína conservada (gene N).
INTRODUCTION
The bovine coronavirus (BCoV) is a pneumoenteric virus classified in group 2 of the Coronaviridae family. The virus consists of an enveloped capsid 120-160 nm in diameter and a helical nucleocapsid of 11-13 nm. The genome is a single-stranded, positive-sense RNA molecule of 27 to 32 kb that encodes 5 major structural proteins, including the nucleocapsid protein (N) and the hemagglutinin-esterase protein (ICTVdB 2009).
In young calves, BCoV is associated with acute diarrhea and, as the morbidity rate can be high, leads to significant economic losses worldwide (Stipp et al., 2009). However, BCoV-induced mortality is usually low (Saif and Heckert, 1990). The virus has also been implicated in winter dysentery of adult cattle and respiratory disease in young cattle (Clark, 1993; Takiuchi et al., 2009).
Several diagnostic techniques including electron microscopy, hemagglutination (HA), hemagglutination inhibition (HI), enzyme-linked immunosorbent assay, and reverse transcription-polymerase chain reaction (RT-PCR) have been used for BCoV detection in stool samples (Sato et al., 1977; Bulgin et al., 1989; Kapil et al., 1999; Schoenthaler and Kapil, 1999). Recently in Brazil, a semi-nested PCR assay was described as a sensitive method for the detection of viral particles in fecal samples from naturally infected calves (Takiuchi et al., 2006).
The isolation of coronaviruses in tissue culture is not routinely used in diagnosis as it requires a relatively long time to be performed. However, one advantage of BCoV adaptation in cell culture is that it can be used for antigenic and genomic characterization, which is indispensable for the development of control measures like vaccines. BCoV has already been propagated in several cell lineages, but it has only been reported once in Brazil and this was in cell line other than HRT-18 (human rectal tumor cells), which is the most sensitive lineage for primary isolation (Jerez et al., 2005). Therefore, the adaptation of wild-type virus strains in cell culture is very useful for antigenic and molecular analysis, particularly for widely spread viruses.
The present study is the first to describe the isolation, in HRT-18 cells, of wild-type Brazilian BCoV from diarrheic stool samples of naturally infected calves. Additionally, the culture-adapted BCoV strains were molecularly characterized.
MATERIALS AND METHODS
Stool samples
Three fecal samples (BCVN1, BCVN2, and BCVN3) taken from calves up to thirty days old with clinical signs of diarrhea, which had previously been confirmed as BCoV-positive using a semi-nested PCR assay, were used in the study. The samples were from calves raised in the Brazilian states of Mato Grosso (MT) and Paraná (PR).
The suspensions for virus isolation in HRT-18 cells were prepared as 20% (w/v) fresh diarrheic fecal sample in 0.01 M phosphate-buffered saline (PBS). They were centrifuged at 3,000 x g for 15 min and stored at 4 ºC until used.
Cells
HRT-18 cells were grown in Dulbecco's Modified Eagle Media (DMEM, Gibco BRL®, USA), supplemented with 10% fetal bovine serum (FBS, Gibco BRL®, USA), 55 mg/mL gentamicin (Sigma Co., USA), and 2.5 mg/mL amphotericin B (Bristol-Meyers Squibb Co., Brazil). Confluent monolayers were washed with calcium- and magnesium-free PBS containing 5 mg/mL trypsin (Sigma Co., USA). The fecal suspensions, which had previously been treated with gentamicin (100 mg/mL) and amphotericin B (10 mg/mL), were then added to the monolayers. The virus was allowed to adsorb at 37 ºC for 90 min in a roller system, and the inoculated HRT-18 cells were kept in a roller drum for 5 days at 37 ºC.
Over the next 3 to 5 days, the monolayers were examined daily for cytopathic effects (CPEs). After three passages from the blind passage, or when approximately 75% of the cells had CPEs, three freeze-thaw cycles were used to release the BCoV, which was then harvested by pooling the cells and supernatants. Non-inoculated HRT-18 cells were included as a negative control.
Hemagglutination assay (HA)
HA was performed using the microtiter method in U-bottom plates (BrandTech Scientific Inc., USA). Serial two-fold dilutions of non-purified, adapted wild-type BCoV cell culture were prepared in 25 mL of 0.01 M PBS, containing 0.1% BSA (Sigma Co., USA), and mixed with 25 mL of a 1.5% mouse erythrocyte suspension. The plates were then incubated at 4 ºC for 2 h. HA titers were expressed as the reciprocal of the highest dilution with complete hemagglutination.
RNA extraction and semi-nested PCR
To confirm virus isolation, 100 mL suspensions from the infected HRT-18 cells were prepared. The suspensions were treated with SDS at a 1% (v/v) final concentration and kept at 56 ºC for 30 min. The RNA extraction was performed by combining the phenol/chloroform/isoamyl alcohol and silica/guanidinum isothiocyanate methods, as described by Alfieri et al. (2006). The same technique was employed to obtain the BCoV RNA from 400 mL of 20% (w/v) fecal suspension. Sterile water was included as a negative control for RNA extraction.
The RT-PCR assay was performed with the primers BCoV1 sense (5'-CGATGAGGCTATTCCGAC-3') and BCoV2 antisense (5'-TGTGGGTGCGAGTTCTGC-3'). BCoV3 sense (5'-TTGCTAGTCTTGTTCTGGC-3') and BCoV2 antisense were used for the semi-nested PCR. The primers were designed by Takiuchi et al. (2006) based on the highly conserved region of the Mebus strain N gene (GenBank accession number U00735) and the technique was performed with methods described in that same study. RT-PCR and semi-nested PCR amplified 454 and 251 bp fragments, respectively. Electrophoresis was performed in a 2% agarose gel stained with 0.5 µg/mL ethidium bromide and visualized under UV light.
RFLP
The specificity of the 251 bp fragments from the semi-nested PCR was confirmed by restriction fragment length polymorphism (RFLP) with the Hae III enzyme (Invitrogen, USA). The reaction was performed following the manufacturer's instructions.
Purification and sequence analysis
The semi-nested PCR amplicons were purified using the QIAquick PCR Purification Kit (Qiagen, USA) and quantified on a 2% agarose gel using Low DNA Mass Ladder (Invitrogen, USA). MegaBACE 1000/Automated 96 Capillary DNA Sequencer, Thermo Sequenase II DNA Polymerase, and the DYEnamic ET Dye Terminator Kit (GE Healthcare, USA) were used for sequencing, which was performed in both directions with forward and reverse primers.
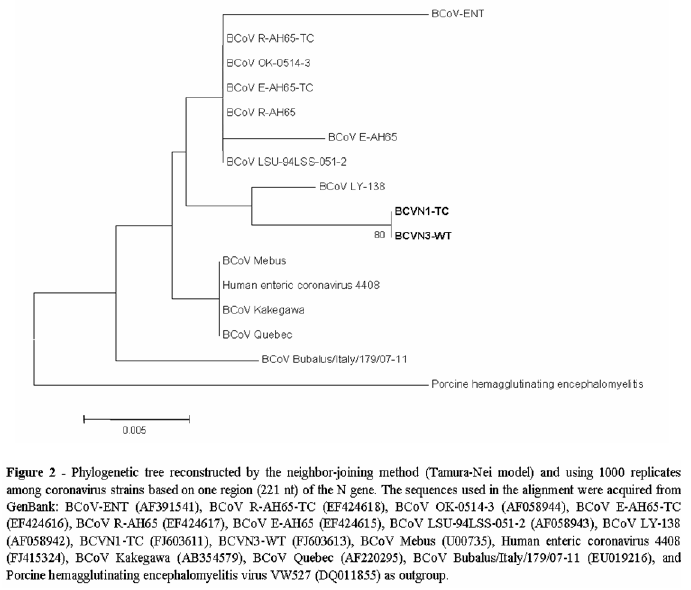
The sequences were analyzed with Sequence Analyzer and Base Caller Cimarron 3.12 software. Sequence quality analysis was performed using Phred and CAP3 software (http://genoma.cenargen.embrapa.br/phph/). Similarity searches were performed with sequences deposited in GenBank using the BLAST software (http://www.ncbi.nlm.nih.gov/ BLAST/). A multiple alignment and an identity matrix were produced in BioEdit software version 7.0.5.3. Phylogenetic tree building was performed using the Neighbor-Joining algorithm based on the Tamura-Nei model, which provided statistical support via bootstrapping with 1,000 replicates using the MEGA package (version 4).
RESULTS
Following three sequential passages in HRT-18 monolayers, viruses from BCVN1 and BCVN2 fecal samples were successfully isolated. Two to three days post-inoculation, CPEs were evident as the cells were granular, swollen, and enlarged. The cell membranes appeared to be fused and resembled syncytia. The cells had focal to diffuse cytoplasmic vacuolation (Fig. 1), especially at four to seven days post-inoculation. Virus isolation from the BCVN3 sample failed. No CPEs were evident in the negative control. BCVN1 and BCVN2 were successfully isolated at titers of 16 and 32 units per 25 µL, respectively (HA assay).
Semi-nested PCR was performed to confirm BCoV isolation and amplified the expected 251 bp fragment of the N gene. The specificity of the products was verified using RFLP with the Hae III enzyme and yielded 88 and 163 bp fragments.
The BCVN1-TC (tissue-culture adapted) and BCVN3-WT (wild-type) partial N gene sequences had 99% nucleotide similarity with each other. The identity with the prototype Mebus was 98.1%. The nucleotide identity ranged from 98.1 to 100% with other published sequences, but BCVN1-TC and BCVN3-WT showed 97.6 to 98.5% identity with the strains included in the study (excluding the out group sequence - Porcine hemagglutinating encephalomyelitis virus). The sequences of BCVN1-TC and BCVN3-WT were deposited in GenBank with the accession numbers FJ603611 and FJ603613. The sequencing of sample BCVN2-TC was unsuccessful.
Phylogenetic analyses placed BCVN1-TC and BCVN3-WT in the same branch with other BCoV strains but separated from the Mebus prototype strain. Although they were clearly in another subgroup, the V270 and LY-138 BCoV strains were less distant from BCVN1-TC and BCVN3-WT (Fig. 2).
DISCUSSION
In an attempt to obtain BCoV from an outbreak of acute diarrhea in calves, three fresh fecal samples were inoculated in HRT-18 cells and two (BCVN1 and BCVN2) isolates were obtained. The success of virus isolation was in part due to the use of a proteolytic enzyme (trypsin), which enhanced the virus infectivity in vitro and activated cell-fusion activity. As with the Kakegawa coronavirus strain, cellular syncytia were observed in inoculated HRT-18 cells. In addition, cytoplasmic vacuolation was also seen, which is in agreement with Benfield and Saif (1990). Isolate specificity was confirmed by each of the other tests performed (i.e., hemagglutination, semi-nested PCR, RFLP, and sequence analysis).
Kapil et al. (1996) isolated BCoV in HRT-18 cells after one passage, and obtained a titer of at least 128 by HA. The relatively low (16 and 32) BCoV HA titers in the present study may be related to the number of passages in HRT-18 cells, as the presence of trypsin in the culture medium can reduce or inhibit the ability of the virus in bind to mouse erythrocytes (hemagglutination), because the surface projections become shorter after two or three passages (Storz et al., 1981).
It has been well established that primary isolation of BCoV in cell culture is difficult, especially for use in routine diagnosis (Kapil et al., 1996). Nevertheless, it is essential to obtain wild-type strains for virological, antigenic, and molecular studies. These isolated BCoV strains may be useful for in vivo pathological studies in biological models (e.g., experimental infection).
The two (BCVN1-TC and BCVN3-WT) semi-nested PCR amplicons sequenced were from two different states, but showed 99% nucleotide identity, indicating that this strain circulates in Brazilian cattle herds from distant geographical areas (MT and PR states). Despite the small size of the N gene fragment analyzed, the identity among Brazilian BCoV strains and the other BCoV strains used in this study were lower (97.6 to 98.5%) than the identity among other published sequences (98.1 to 100%) (data not shown). Since the region of the N gene amplified by the semi-nested PCR was highly conserved among coronaviruses, it is possible that other genes also present low identity; this has previously been observed in the S gene from other Brazilian BCoV strains (Takiuchi et al., 2007; 2008). Thus, additional studies are necessary for molecular characterization of Brazilian BCoV strains. Obtaining viral progeny will be easier, as the virus isolation has already been done, and future studies could focus on the antigenic features of these strains.
It is well-known that multiple virus passages can result in mutation of the viral genome, which would increase the molecular distance between the wild-type and tissue culture strain. Therefore, it is particularly notable that this study used a low number of virus passages in cell culture, as mutations could alter the antigenic features and interfere with strain characterization.
In the dendrogram, BCVN1-TC and BCVN3-WT clustered into the same group, but they were clearly in a different cluster than the main strain, Mebus. Aside from the strain groupings with other BCoV strains, the molecular heterogeneity in this fragment of the N gene, which is supposedly more conserved than the other genes, suggests that there is high genetic variability in the Brazilian BCoV strains obtained in this study.
In conclusion, the present study is the first to describe the isolation of Brazilian BCoV strains in HRT-18 cells. The isolation of these strains is extremely important, as the frequency of coronavirus infection in Brazilian cattle herds is relatively high and future control measures may be necessary. Additionally, the molecular identification of these strains revealed the circulation of a BCoV strain that is divergent from the main BCoV strains that are circulating worldwide.
ACKNOWLEDGEMENTS
The financial resources for the conduction of this study were supported by the project BioAgroPar financed by FINEP, SETI/PR, and Fundação Araucária/PR; and by CNPq/Brazil.
Part of the research activities of this study was carried out in the Agricultural Research Support Laboratory (Laboratório de Apoio à Pesquisa Agropecuária - LAPA) / PROPPG / UEL.
Alfieri, A.A. and Alfieri, A.F. are recipients of CNPq fellowships.
- Alfieri, A. A., Parazzi, M. E., Takiuchi, E., Médici, K. C., Alfieri, A. F. (2006), Frequency of group A rotavirus in diarrhoeic calves in Brazilian cattle herds, 1998-2002. Trop Anim Health Prod, 38, 521-526
- Benfield, D. A., Saif, L. J. (1990), Cell culture propagation of a coronavirus isolated from cows with winter dysentery. J Clin Microbiol, 28, 1454-1457
- Bulgin, M. S., Ward, A. C., Barrett, D. P., Lane, V. M. (1989), Detection of rotavirus and coronavirus shedding in two beef cow herds in Idaho, Can Vet J, 30, 235239
- Clark, M. A. (1993), Bovine coronavirus. Br Vet J, 149, 51-70
- ICTVdB – The Universal Virus Database, version 4. <http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdb/>. Access in: 10 jan. 2009
- Jerez, J. A., Gregori, F., Brandăo, P. E., Rodriguez, C. A. R., Ito, F. H., Buzinaro, M. G., Sakai, T. (2005), Isolation of bovine coronavirus (BCoV) in monolayers of HmLu-1 cells. Braz J Microbiol, 36, 207-210
- Kapil, S., Richardson, K. L., Radi, C., Chard-Bergstrom, C. (1996), Factors affecting isolation and propagation of bovine coronavirus in human rectal tumor-18 cell line. J Vet Diagn Invest, 8, 96-9
- Kapil, S., Richardson, K. L., Maag, T. R., Goyal, S. M. (1999), Characterization of bovine coronavirus isolates/from eight different states in the USA. Vet Microbiol, 67, 221-30
- Saif, L. J., Heckert, R. (1990), Enteropathogenic coronaviruses, In-L. J. Saif and K. W. Theil. (ed.), Viral diarrheas of man and animals CRC Press, Boca Raton, Florida. pp. 185252
- Sato, K., Inaba, Y., Kurogi, H., Takahashi, E., Satoda, K., Omori T., Matsumoto, M. (1977), Hemagglutination by calf diarrhea coronavirus, Vet Microbiol, 2, 8387
- Schoenthaler, S. L., Kapil, S. (1999), Development and application of a bovine coronavirus antigen detection enzyme-linked immunosorbent assay. Clin Diagn Lab Immunol, 6, 130132
- Stipp, D. T., Barry, A. F., Alfieri, A. F., Takiuchi, E., Amude, A. M., Alfieri, A. A. (2009), Frequency of BCoV detection by a semi-nested PCR assay in faeces of calves from Brazilian cattle herds. Trop Anim Health Prod, in press. doi: 10.1007/s11250-009-9347-2
- Storz, J., Rott, R., Kaluza, G. (1981), Enhancement of plaque formation and cell fusion of an enteropathogenic coronavirus by trypsin treatment. Infect Immun, 31, 12141222
- Takiuchi, E., Stipp, D. T., Alfieri, A. F., Alfieri A. A. (2006), Improved detection of bovine coronavirus N gene in faeces of calves infected naturally by a semi-nested PCR assay and an internal control. J Virol Methods, 131, 148-154
- Takiuchi, E., Barreiros, M. A. B., Alfieri, A. F., Alfieri, A. A. (2007), Identification of a mutation in the spike protein cleavage site in Brazilian strains of wild-type bovine coronavirus. Braz J Microbiol, 38, 699-703
- Takiuchi, E., Alfieri, A. F., Alfieri, A. A. (2008), Molecular analysis of the bovine coronavirus S1 gene by direct sequencing of diarrheic fecal specimens. Braz J Med Biol Res, 41, 277-282
- Takiuchi, E., Barry, A. F., Alfieri, A. F., Filippsen, P., Alfieri, A. A. (2009), An Outbreak of Winter Dysentery Caused by Bovine Coronavirus in a High-Production Dairy Cattle Herd from a Tropical Country. Braz Arch Biol Technol, in press
Publication Dates
-
Publication in this collection
11 Feb 2010 -
Date of issue
Nov 2009