ABSTRACT
The present investigation details an assessment of genetic relationship of E. coli isolates collected from two different environmental sources viz. sewage water and soiled bedding materials of laboratory rodents. Five sewage water samples were collected from the industrial area of Lucknow city and 5 samples of soiled bedding materials of laboratory animals were collected from Animal facility at CSIR-IITR, Lucknow. For this study Random amplified polymorphic DNA markers (RAPD) was chosen as the molecular fingerprinting method. In this study, 10 RAPD primers were used to evaluate the genetic similarity of E. coli. isolates. The RAPD-PCR fingerprints were analyzed and data were scored as 1, 0 matrix. The generated data were fed on Popgene software for calculating genetic diversity and creating dendrogram. The genetic similarity of 85% was recorded from soiled bedding materials and only 71% in sewage water samples in E.coli samples. The dendrogram based generation of clustering of E. coli isolates show two major clusters. Within major cluster sub-cluster were also observed which indicating diversity within isolates of E. coli. The RAPD-PCR based fingerprinting provided a rapid means of discriminating E. coli isolates and considered a relevant tool for molecular typing.
Key words:
Escherichia coli; Genetic similarity; RAPD-PCR and Rodents
INTRODUCTION
The Escherichia coli have been intensively studied under various aspects in general bacteriology. E.coli is a common inhabitant of the gastrointestinal tract of most animals, including birds, but not all E.coli isolates are capable of causing diseases [1[1] Nataro J.P, Kaper J.B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998; 11: 142 -201.]. Analysis of E.coli strains was isolated from chicken by Random Amplified DNA marker [2[2] Kilic, A, A. H. Muz and G. Ertas. Random Amplified Polymorphic DNA (RAPD) Analysis of Escherichia coli Isolated From Chickens. F.Ü. Sag. Bil. Vet. Derg. 2009; 23 (1):01-04.]. The pathogenic Escherichia coli, gram negative bacteria, have been widely studied under different biomedical research like vehicles for the cloning of genes, testing of efficacy of antimicrobial agents, indicator organisms, etc. DNA fingerprinting of the species of Escherichia and Shigella revealed that E. coli is very closely related to Shigella [3[3] Cui S, Schroeder C M, Zhang D Y & Meng J. Rapid sample preparation method for PCR-based detection of Escherichia coli 0157: H7 in ground beef, J. Appl. Microbiol. 2003; 95(1):129-34.]. The bacteriological examination of water has a special significance in pollution studies, as it is a direct measurement of deleterious effect of pollution on human health [4[4] Singh C., Singh J S., et al. Screening out of coliform bacteria from different location of Gomati River in Lucknow. African Journal of Microbiology. 2013; 7(29), 3762-3771.]. Differentiation of Escherichia coli strains using randomly amplified polymorphic DNA analysis and protein biochemical markers were studies [5[5] Wang G, Whittam T S, Berg C M & Berg D E. RAPD (arbitrary primer) PCR is more sensitive than multi locus enzyme electrophoresis for distinguishing related bacterial strains, Nucleic Acids Res, 1993; 21,5930-5933.]. Identification and differentiation of E.coli strains were isolated from clinical samples by RAPD marker [6[6] Cave H, Bingen E, Elion J & Denamur E. Differentiation of Escherichia coli strains using randomly amplified polymorphic DNA analysis, Res Microbiol. 1994; 145, 141-150.]. The other genotyping studies carried out on clinical, environmental and veterinary isolates, using E. coli previously identified via biochemical and/or culture based approaches [7[7] Gordon D.M. The genetic structure of Escherichia coli populations in feral house mice Microbiology.1997; 143, 2039 - 2046.]. Isolation and identification of Escherichia coli O157:H7 using different detection methods and molecular determination by multiplex PCR and RAPD [8[8] Kim J.Y., Kim S.-H., Kwon N.-H., et al., Isolation and identification of Escherichia coli O157:H7 using different detection methods and molecular determination by multiplex PCR and RAPD, Journal of Veterinary Science.2005; 6, 7-19,]. Using of multiplex PCR for Detection of Escherichia coli O157:H7 and analysis of RAPD, PFGE, plasmid profiling and antimicrobial property [9[9] Radu S., Ling O. W., Rusul G., Karim M. I. A., Nishibuchi M. Detection of Escherichia coli O157:H7 by multiplex PCR and their characterization by plasmid profiling, antimicrobial resistance, RAPD and PFGE analyses,” Journal of Microbiological Methods. 2001; 46, 131-139. International Journal of Food Microbiology. 2003; 84(1) 63- 69.]. Isolation of Escherichia coli O157 strain and their molecular characterization isolated from cattle, pigs and chickens at slaughter [10[10] Kumar M., Kumar S. Genetic structure and inter-generic relationship of closed colony of laboratory rodents based on RAPD markers, Mol. Biol. Rep. 014; 41(11):7273-80.]. In our previous study the evaluation of genetic analysis of rodents performed by RAPD techniques. [11[11] Tseng, C. C., W. T. E. Ting, Y. Cheng, D. Johnson and M. Saluta. A comparative study of RAPD fingerprints of Escherichia coli isolates from humans and animals. Abstract Q-69, p.546. In Abstracts of the 99th General Meeting of the American Society for Microbiology 1999. American Society for Microbiology. Washington, D.C.]. The RAPD analysis uses single primers to amplify the genomic DNA as compare to restriction fragment length polymorphism (RFLP). Each primer can generate a specific DNA profile per sample [2[2] Kilic, A, A. H. Muz and G. Ertas. Random Amplified Polymorphic DNA (RAPD) Analysis of Escherichia coli Isolated From Chickens. F.Ü. Sag. Bil. Vet. Derg. 2009; 23 (1):01-04.]. Genetic characterization of Escherichia coli isolates from humans and animals sample by RAPD marker [12[12] Vogel, L., Oorschot E. Van, Maas H. M. E., Minderhoud B., Dijkshoorn L. Epidemiologic typing of Escherichia coli using RAPD analysis, ribotyping and serotyping Clinical Microbiology and Infection. 6:82-87. feral house mice, Microbiology. 2000; 143, 2039-2046.]. The RAPD analysis, ribotyping and serotyping of E.coli an epidemiological study was carried by Vogel et al., (2000) [13[13] Maity B and Guru P Y. Genetic diversity and molecular characterization of pathogenic Escherichia coli from different species of laboratory rodents, Indian Journal of Biotechnology 2006; 6, 210-215.]. Some studies were also done in isolation of E.coli from house mice (wild mice) [7[7] Gordon D.M. The genetic structure of Escherichia coli populations in feral house mice Microbiology.1997; 143, 2039 - 2046.]. Some studies in isolated and screening of E.coli from different location in Gomati river Lucknow city have also been conducted [4[4] Singh C., Singh J S., et al. Screening out of coliform bacteria from different location of Gomati River in Lucknow. African Journal of Microbiology. 2013; 7(29), 3762-3771.]. Few studies have been conducted on genetic diversity of pathogenicEscherichia coli from different species of laboratory rodents by RAPD marker [14[14] Mishra S.K., Kumar M., Sharma R.P., Rajnarayan, Tyagi J.S. Assessment of generic differences between and within four avian species by employing randomly amplified polymorphic DNA markers.Indian Journal of Poultry Science 2008; 43:257-261.]. Ultimately there are few studies done on to the pathogenicity of E.coli strains from industrial site of sewage water sample and compared with laboratory animals (rodents) soil bedding material because the sewage water directly falls into Gomati river and contaminates the water of Gomati river which causes different diseases to among humans as well as animals. The Gomati river is an important river which passes from Lucknow city and is also a main source of drinking water supply. Gomti river receives huge quantities of untreated sewage agricultural runoff which brings lot of pesticides, pathogen microbes especially of E.coli. Shigella and salmonella species; industrial wastes significantly alter the physicochemical characteristics of its water. Very few studies has been conducted and there is scant literature on phylogenetic relationship of E. coli strains in bedding material of different laboratory rodents and sewage water from Gomati river [7]. While monitoring of rodents from animal house facility and Gomati River, the pathogenic E.coli strains play an important role in colony management of Gomati river and laboratory animal facility.
The aim of this study was to evaluate the genetic diversity of E.coli from two different Environmental Sources, Therefore 10 RAPD primers were used for the genetic diversity and molecular characterization of E.coli strains from two different places in Lucknow city India.
The RAPD technique provides a simple, fast and a comparatively low-cost marker system which has gained wide acceptance, world-wide [15[15] Sambrook J, Russell DW. Molecular cloning a laboratory manual, 3rd edn. Cold Spring Harbor laboratory Press, New York; 1989.]. In the present investigation of this study was to evaluate the genetic analysis of Escherichia coli isolated from two different environmental Sources: Sewage Water Verses Soiled Bedding Materials of Laboratory Rodents based on RAPD markers.
MATERIALS AND METHOD
Bacterial culture and growth condition
The present study was carried out at Environmental Microbiology Section CSIR-Indian Institute of Toxicology Research Lucknow India. The samples are collected from possible distinct environmental conditions. Five water samples and five bedding samples were taken initially for analysis of E.coli strains: such as from sewage area near Gomati River and animal facility room in IITR Lucknow.
In media preparation the nutrient broth was prepared by dissolving 13 gram of dehydrated nutrient broth (Himedia, India) into 1000 ml of distilled water and was sterilized by autoclaving at 1210C under 15l b pressure per square inch for 15 minutes. Then the broth was dispensed into tubes (10 ml/tube) and stored at 40C in the refrigerator until used. The protocol for growing the E.coli bacteria was followed with Red hot sterilized platinum loop bein used to streak the nutrient broth culture on EMB agar or MacConkey agar for isolating colony. The media containing streaked culture was kept at 370C for overnight in incubator. The colony of pink/red and greenish black with metallic sheen appears on MacConkey and EMB agar plates respectively.
Biochemical Identification
There are different biochemical test to use to identify the E.coli bacterial strains like-
Catalase Test: Using a glass capillary tube, small amount of culture from the plate was used. Care was taken to ensure that no blood strains were present as the presence of catalase in the medium itself may give a false positive result. To the observed the immediate formation of oxygen bubbles in the tube indicating the activity of catalase. Each culture was observed for the appearance of or absence of gas bubbles.
Iodole Test: Bacterium to be tested is inoculated in peptone water, which contains amino acid tryptophan and incubated overnight at 370C. After incubation few drops of Kovac’s reagent were added, Kovac’s reagent consists of para-dimethyl aminobenzaldehyde, isoamyl alcohol and conc. HCL. Ehrlich’s reagent is more sensitive in detecting indole production in anaerobes and non-fermenters. The tube color was examined in the reagent “layer”.
Methyl Red (MR) Test: The bacterium to be tested in inoculated into glucose phosphate broth, which contains glucose and phosphate buffer and incubated at 370C for 48 hours. Over the 48 hours the mixed acid producing organism should produce sufficient acid to overcome the phosphate buffer and remaining acid. The pH of the medium is tested by the addition of 5 drops of MR reagent. The change in color of methyl red or MR test was observed.
Voge’sProskauer (VP) Test: Bacterium to be tested is inoculated into glucose phosphate broth and incubated for at least 48 hours. 0.6 ml of alpha-naphthol is added to the test broth and shaken. 0.2 ml of 40% KOH is added to the broth and shaken. The tube is allowed to stand for 15 minutes. The negative tubes must be held for one hour, since maximum color development occurs within one hour after addition of reagents. Observe the tubes for change in color for the VP test.
Citrate Utilization Test: Preparation of Simmon’s citrate agar slants. Inoculate Simmon’s citrate agar slants, by means of stab-and-streak inoculation. The one tube is kept as an un inoculated comparative control. Incubate all the three slants at 370C for 48 hours. Observe the slant cultures for the growth and coloration of the medium.
Gram’s stain: In gram’s staining under microscope the organism revealed gram-negative, pink color, small rod shaped, single or paired. The representative figure of Gram’s staining is as follows.
Identification and DNA extraction of E.coli strains
After biochemically and staining processes, 10 samples were identified as containing E.coli colonies than confirmed pathogenic E. coli species were isolated from sewage water sample and bedding material of laboratory animal species. DNA was extracted from E. coli after inoculation in Luria broth followed by incubation at 37°C in tail digestion buffer and 20% SDS buffer with 5 µl proteinase K followed by phenol: chloroform: isoamyl extraction and ethanol precipitation with 3 M Sodium acetate according to Sambrook and Russell (1989)[16] protocol. Ten samples (five from sewage water sample and five from bedding material) from each sources were screened for E.coli isolate and subsequently DNA was extracted and stored at -20 oC for future use.The quantity of genomic DNA was estimated by spectrophotomer and DNA samples loaded on 0.8 % agarose gel.
RAPD-PCR
A total of 15decamer primers from the set OPA, OPG and OPO (Operon Biotechnologies GmbH, Germany) having GC content 60-70 % were initially screened RAPD PCR. The oligonucleotide primers information along with amplified band size range and GC content is provided in Table 1.The RAPD-PCR reactions were performed in a mixture of 25 µl volume containing 50 ng of genomic DNA , 12.5 µl hot start master mix (Biotool)), 1.0µL (25 pmole) primer and volume make up nuclease free water. DNA was amplified by a thermal circler (mycycler, BioRadTM, USA) using the following cycling conditions: first denaturation of 5 min at 94°C followed by 45 cycles of 1 min at 94°C, 1 min at 35°C and 1 min at 72°C and final extension at 72°C for 5 min. One negative control was used in each primer reaction. The PCR reactions were carried out as described by Maity and Guru (2006) [14[14] Mishra S.K., Kumar M., Sharma R.P., Rajnarayan, Tyagi J.S. Assessment of generic differences between and within four avian species by employing randomly amplified polymorphic DNA markers.Indian Journal of Poultry Science 2008; 43:257-261.]. The PCR amplified products were resolved on 1.75 % agarose gel having ethidium bromide in 1X TBE buffer [45 mMTris-borate, 1.0 mM EDTA (pH 8.0)] at 75 V for 4 -5 h . A low range ladder was used for molecular weight determination. The amplified band pattern were visualized and photographed in a gel documentation system using Gene snap software (Syngene), UK.
Primer name, Sequence, GC content, Tm value oC, Size of amplicons and Number of bands scored.
RAPD data analysis
The analysis of allele size amplified by different RAPD markers was done in Genesnap and Gene tool softwares (Syngene, UK) along with the standard size ladders (100 and 500 bp).After matching the bands pattern in different gels than bands were scored as ‘1’ and ‘0’ for their presence and absence respectively. For molecular genetic investigation the binary coded characters (1, 0) were used. From generated binary data, the level of polymorphism was analysed by dividing the polymorphic bands by the total number of scored bands. Similarly, % of polymorphic loci was calculated for each genus using popgene version 1.31 software [17[17] Higgins J., Hohn C., Hornor S., (2007) Genotyping of Escherichia coli from environmental and animal samples, Journal of Microbiological Methods. 2007; 70, 227-235.].
RESULTS
On the basis of culture examination and biochemical parameter of isolated bacteria from Gomati sewage water and soiled bedding material of laboratory animals are given bellow.
Biochemical test
The samples were collected and promptly inoculated on McConkey agar plate. After overnight incubation, bright pink colonies were observed in 10 Gomati River and soil bedding material samples. The presumptive colony on McConkey agar for each bedding material was sub cultured successively onto Eosin Methylene Blue (EMB) agar for presumptive identification of E. coli. The greenish-black colonies with metallic sheen on EMB agar were observed. In gram’s staining under microscope the organism revealed gram-negative, pink color, small rod shaped, single or paired. Presumptively selected 2 to 3 mm colonies were repeatedly streaked on the respective selected media (EMB agar) to check and confirm their purity. For identification, a series of biochemical tests especially selective for E. coli were performed with the culture positive and gram-negative rod shaped cells. All the isolates are fermented the two sugars producing acid and gas. Acid production is indicated by the color change from reddish to yellow and the gas production was noted by the appearance of gas bubbles in the inverted Durham’s tubes.
PCR product analysis
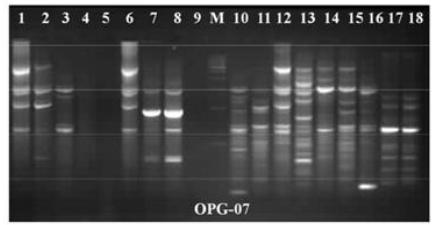
In this study, 20 RAPD primers for amplification of bacterial genomic DNA were selected from two different sources. Out of 20 Only 12 primers formed the best, suitable, clear, and reproducible banding patterns. The amplified band patterns were found to be clear and reproducible when reaction were repeated using the same reaction condition. The total 165 types of bands were amplified by 12 RAPD primers i.e. scorable and easily counted. All 20 RAPD primers yielded reproducible DNA profiles. Control assays in which distilled water was used in place of template yielded no amplified products. The polymorphism was counted in different E.coli strains from different sources, some bands similar and some bands dissimilar reported. The primer OPG-02, OPG-07 and OPG -14 produced good results for investigation of polymorphism in all strains. The primer OPG-02 and OPG-07 approximately 250 bp and 600 bp allele size were similar in all strains of E.coli. (Fig. 1).The primer OPG-12 and OPG-07 produced good polymorphism in all strains of E.coli bacteria (Fig. 2&4). The numbers of scored bands, band size ranges were represented in Table 2. The molecular weight of amplified bands ranged from 200 bp to 3.4kbp. The maximum size-range of amplified products (230- 2,400 bp) for a single primer was found with OPG- 02, whereas the minimum (150-1,900 bp) was obtained with OPG-07. The maximum number of bands (11) was produced with primer OPG-02 (Fig. 3) and the minimum (1) with primer OPG-06. The RAPD DNA finger printing with 20 primers generated a total of 2,120 bands in all samples.
RAPD-PCR gel patterns of primer OPG-02 showing E. coli samples. Lane M is 100 bp molecular weight marker ladder, lane 1-4 E.coli from Gomati river samples and lane 5- 8 E.coli from soil bedding material samples.
RAPD-PCR gel patterns of primer OPG-12 showing E.coli samples. Lane M is 50 bp molecular weight marker ladder, lane 1-5 E.coli from Gomati river samples and lane 6- 10 E.coli from soil bedding material samples.
RAPD-PCR gel patterns of primer OPG-02 showing E.coli samples. Lane M is 100 bp and 1 kbp molecular weight marker ladder, lane 1-10 E.coli from Gomati river samples and lane 11-119 E.coli from soil bedding material samples.
RAPD-PCR gel patterns of primer OPG-07 showing E.coli samples. Lane M is 100 bp molecular weight marker ladder, lane 1-9 E.coli from Gomati river samples and lane 10 -18 E.coli from soil bedding material samples.
Phylogenetic tree analysis
In the dendrogram analysis of all E.coli Strains are grouped into two major clusters A and B. The cluster A contains E.coli strain isolated from Gomati River and cluster B contain soil bedding material of laboratory animals. The cluster A further separated in five different sub clusters isolated from Gomati River likewise cluster B separated also separated in five different sub culture isolated from soil bedding material from laboratory animals (Fig. 5). The genetic similarity of 85% was seen in E. coli samples isolated from soiled bedding materials and only 71 % were observed in sewage water samples .The Nei’s genetic similarities and genetic differences were among all E.coli Strains were given in Table 3.
Genetic similarity (above diagonal) genetic distance (below diagonal) Sample 1-5 E.coli isolated from sewage water and 6-10 soil bedding material of laboratory rodents.
DISCUSSION
In our study, the genetic diversity of E.coli strains was investigated by RAPD PCR techniques. Evaluation of genetic diversity among different E.coli strains at genomic level was analysed by RAPD PCR technique [18[18] Gomes A.R., Muniyappa L., Krishnappa G., et al. Genotypic Characterization of Avian Escherichia coli by Random Amplification of Polymorphic DNA. Int. Journal of poultry science 2005; 4 (6): 378-381,-19[19] Muzurier, S.I., Wernas K. Typing of Listeria strains by random amplification of polymorphic DNA. Res. Microbiol.1992; 143: 499-505.]. Several arbitrary primer based RAPD-PCR technique has been used for delineating the bacteria according to their genetic relatedness [20[20] Eisen, D., E.G. Russel, M. Tymms, E.J. Roper, M.L. Grayson and J. Turnidge, Random amplified polymorphic DNA and plasmid analysis used in investigation of an outbreak of multi resistantKlebsiella pneumoniae. J. Clin. Microbiol.1995; 33: 713-717.
[21] Lin, A.W., M.A. Usera., T.J. Barrettand and R.A. Goldsby. Application of random amplified polymorphic DNA analysis to differentiate strains of Salmonellae enteritidis. J. Clin. Microbiol. 1996; 4: 870-876.-22[22] Johnson J R, O’ Bryan T T, Low D A, Ling G, Delavari P et al. Evidence of commonality between canine and human extra intestinal pathogenic Escherichia coli that express papGallele III, Infect Immun,2000; 68, 3327-3336.]. In this study reveals with DNA marker RAPD-PCR based genetic analysis of different E.coli strains was compare with difference place of other E.coli strains [14[14] Mishra S.K., Kumar M., Sharma R.P., Rajnarayan, Tyagi J.S. Assessment of generic differences between and within four avian species by employing randomly amplified polymorphic DNA markers.Indian Journal of Poultry Science 2008; 43:257-261.]. In previous year there is no evidential data were found to be differentiate to the E.coli strain in different region using RAPD technique. In this study we monitored the Gomati River in Lucknow city and bedding material Laboratory animal. However, we believe that the genetic analysis of E.coli based on PCR method evaluate in this paper, may offer a cost-effective and time saving of E. coli isolates from different type of sample. The amplified fragment size of E.coli strain was investigated to the band size average between 200 bp to 3.4 kbp. The E.coli is the most important species in the genus Escherichia and recognized as an important potential pathogenic in humans as well as animals [23[23] Potard A B B, Colin T, Scaletsky I, Bougunec C L, Guignot J et al. Representational difference analysis between Afa/Dr diffusely adhering Escherichia coli and non pathogenic Escherichia coli K-12, Infect Immun, 2002; 70, 5503-5511.]. The other types of study were carried out in some animal species such as Identification and differentiation of pathogenic E. coli strains in animals using RAPD-PCR, multiplex-PCR [24[24] Clermont O, Bonacorsi S &Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group, Appl Environ Microbiol. 2000; 66, 4555-4558.-26]. The Result showed that the significant effect of E.coli in different places such information indicate that the Gomati river isolates will be more diversity when compare to soil bedding material isolates. The primerOPA-12 ( Fig. 3) having GC content 70 % produces highest number of bands in all the E.coli strains. The results agreements with other types of study of E.coli in laboratory rodents were investigated [14[14] Mishra S.K., Kumar M., Sharma R.P., Rajnarayan, Tyagi J.S. Assessment of generic differences between and within four avian species by employing randomly amplified polymorphic DNA markers.Indian Journal of Poultry Science 2008; 43:257-261.]. The most challenging aspect of a protocol for E.coli diversity by RAPD study was the selection of suitable primers. We found that primers OPO 12, OPA 12, OPA13, OPG 01, OPG 07 and OPG 11 generated totally different banding pattern for each E.coli and could differentiate all the strains from one another. A phylogenetic tree was constructed by popgene software in all E.coli samples based on RAPD marker. These dendrogram generated from different primers were separated from one another, suggestive of genetically different patterns in different places. The similar type of study was also reported by [14[14] Mishra S.K., Kumar M., Sharma R.P., Rajnarayan, Tyagi J.S. Assessment of generic differences between and within four avian species by employing randomly amplified polymorphic DNA markers.Indian Journal of Poultry Science 2008; 43:257-261.]. The genetic similarities and genetic distance was found to be more informative to know the diversity of E.coli isolated from two different places. From this of study, it is concluded that RAPD markers were useful in detecting, genetic similarities, genetic diversities and genetic distances in different pathogenic of E.coli strain in different places.
These results also help to the understanding of the working on E. coli species in laboratory rodents in biomedical research. Thus, we suggest that application of this type of study in molecular characterization of E.coli infections to understand the pathogenic and diversity of the bacteria isolated in Gomati river and bedding material of laboratory animals.
CONCLUSION
The E. coli strains causing diarrhoea in different laboratory animal species were characterized using RAPD primers. There are very few variation in E. coli strains among the animal species suffering from diarrhoea has been found. Identification of pathogenic E. coli strains causing diarrhoea in laboratory animal species was observed by biochemical analysis of pathogenic E.coli. The primer designing are also required for quick identification of pathogenic gene of E. coli species in laboratory animals for further studies.
ACKNOWLEDGMENTS
Authors are grateful to Director, CSIR-Indian Institute of Toxicology Research, Lucknow for the support and encouragement. The authors acknowledge Council of scientific and Industrial Research (CSIR), Government of India, New Delhi, for financial assistance under empower project. The corresponding author is also grateful to Head, Amity Institute of Biotechnology for providing the lab facility to conducting the research work.
REFERENCES
-
[1]Nataro J.P, Kaper J.B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998; 11: 142 -201.
-
[2]Kilic, A, A. H. Muz and G. Ertas. Random Amplified Polymorphic DNA (RAPD) Analysis of Escherichia coli Isolated From Chickens. F.Ü. Sag. Bil. Vet. Derg. 2009; 23 (1):01-04.
-
[3]Cui S, Schroeder C M, Zhang D Y & Meng J. Rapid sample preparation method for PCR-based detection of Escherichia coli 0157: H7 in ground beef, J. Appl. Microbiol. 2003; 95(1):129-34.
-
[4]Singh C., Singh J S., et al. Screening out of coliform bacteria from different location of Gomati River in Lucknow. African Journal of Microbiology. 2013; 7(29), 3762-3771.
-
[5]Wang G, Whittam T S, Berg C M & Berg D E. RAPD (arbitrary primer) PCR is more sensitive than multi locus enzyme electrophoresis for distinguishing related bacterial strains, Nucleic Acids Res, 1993; 21,5930-5933.
-
[6]Cave H, Bingen E, Elion J & Denamur E. Differentiation of Escherichia coli strains using randomly amplified polymorphic DNA analysis, Res Microbiol. 1994; 145, 141-150.
-
[7]Gordon D.M. The genetic structure of Escherichia coli populations in feral house mice Microbiology.1997; 143, 2039 - 2046.
-
[8]Kim J.Y., Kim S.-H., Kwon N.-H., et al., Isolation and identification of Escherichia coli O157:H7 using different detection methods and molecular determination by multiplex PCR and RAPD, Journal of Veterinary Science.2005; 6, 7-19,
-
[9]Radu S., Ling O. W., Rusul G., Karim M. I. A., Nishibuchi M. Detection of Escherichia coli O157:H7 by multiplex PCR and their characterization by plasmid profiling, antimicrobial resistance, RAPD and PFGE analyses,” Journal of Microbiological Methods. 2001; 46, 131-139. International Journal of Food Microbiology. 2003; 84(1) 63- 69.
-
[10]Kumar M., Kumar S. Genetic structure and inter-generic relationship of closed colony of laboratory rodents based on RAPD markers, Mol. Biol. Rep. 014; 41(11):7273-80.
-
[11]Tseng, C. C., W. T. E. Ting, Y. Cheng, D. Johnson and M. Saluta. A comparative study of RAPD fingerprints of Escherichia coli isolates from humans and animals. Abstract Q-69, p.546. In Abstracts of the 99th General Meeting of the American Society for Microbiology 1999. American Society for Microbiology. Washington, D.C.
-
[12]Vogel, L., Oorschot E. Van, Maas H. M. E., Minderhoud B., Dijkshoorn L. Epidemiologic typing of Escherichia coli using RAPD analysis, ribotyping and serotyping Clinical Microbiology and Infection. 6:82-87. feral house mice, Microbiology. 2000; 143, 2039-2046.
-
[13]Maity B and Guru P Y. Genetic diversity and molecular characterization of pathogenic Escherichia coli from different species of laboratory rodents, Indian Journal of Biotechnology 2006; 6, 210-215.
-
[14]Mishra S.K., Kumar M., Sharma R.P., Rajnarayan, Tyagi J.S. Assessment of generic differences between and within four avian species by employing randomly amplified polymorphic DNA markers.Indian Journal of Poultry Science 2008; 43:257-261.
-
[15]Sambrook J, Russell DW. Molecular cloning a laboratory manual, 3rd edn. Cold Spring Harbor laboratory Press, New York; 1989.
-
[16]Yeh F.C, Yang R.C, Boyle T. Popgen version 1.32, a Microsoft window-based freeware for population genetic analysis, quick user guide. Center for International Forestry Research, University of Alberta, Edmonton, Alberta, Canada 1999.
-
[17]Higgins J., Hohn C., Hornor S., (2007) Genotyping of Escherichia coli from environmental and animal samples, Journal of Microbiological Methods. 2007; 70, 227-235.
-
[18]Gomes A.R., Muniyappa L., Krishnappa G., et al. Genotypic Characterization of Avian Escherichia coli by Random Amplification of Polymorphic DNA. Int. Journal of poultry science 2005; 4 (6): 378-381,
-
[19]Muzurier, S.I., Wernas K. Typing of Listeria strains by random amplification of polymorphic DNA. Res. Microbiol.1992; 143: 499-505.
-
[20]Eisen, D., E.G. Russel, M. Tymms, E.J. Roper, M.L. Grayson and J. Turnidge, Random amplified polymorphic DNA and plasmid analysis used in investigation of an outbreak of multi resistantKlebsiella pneumoniae. J. Clin. Microbiol.1995; 33: 713-717.
-
[21]Lin, A.W., M.A. Usera., T.J. Barrettand and R.A. Goldsby. Application of random amplified polymorphic DNA analysis to differentiate strains of Salmonellae enteritidis. J. Clin. Microbiol. 1996; 4: 870-876.
-
[22]Johnson J R, O’ Bryan T T, Low D A, Ling G, Delavari P et al. Evidence of commonality between canine and human extra intestinal pathogenic Escherichia coli that express papGallele III, Infect Immun,2000; 68, 3327-3336.
-
[23]Potard A B B, Colin T, Scaletsky I, Bougunec C L, Guignot J et al. Representational difference analysis between Afa/Dr diffusely adhering Escherichia coli and non pathogenic Escherichia coli K-12, Infect Immun, 2002; 70, 5503-5511.
-
[24]Clermont O, Bonacorsi S &Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group, Appl Environ Microbiol. 2000; 66, 4555-4558.
-
[25]Reid S D, Betting D J &Whittam T S. Molecular detection and identification of intimin alleles in pathogenic Escherichia coli by multiplex PCR, J Clin Microbiol, 1999; 37, 2719-2722.
Publication Dates
-
Publication in this collection
2017
History
-
Received
03 Feb 2016 -
Accepted
14 July 2016