Abstract
Bacterial contamination of blood components remains a major challenge in transfusion medicine, particularly, platelet concentrates (PCs) due to the storage conditions that support bacterial proliferation. In this study, we develop a rapid, sensitive and specific real-time PCR protocol for bacterial screening of PCs. An internally controlled real-time PCR-based method was optimized and validated with our proprietary 16S Universal PCR Master Mix (IBMP/Fiocruz), which targets a conserved region of the bacterial 16S rRNA gene. Nonspecific background DNA was completely eliminated by treating the PCR Master Mix with ethidium monoazide (EMA). A lower limit of detection was observed for 10 genome equivalents with an observed Ct value of 34±1.07 in calibration curve generated with 10-fold serial dilutions of E. coli DNA. The turnaround time for processing, including microbial DNA purification, was approximately 4 hours. The developed method showed a high sensitivity with no non-specific amplification and a lower time-to-detection than traditional microbiological methods, demonstrating it to be an efficient means of screening pre-transfusion PCs.
Keywords:
bacterial contamination; real-time PCR; molecular testing; platelet concentrates
Resumo
A contaminação bacteriana dos componentes sanguíneos é um grande desafio na medicina transfusional, principalmente nos concentrados de plaquetas (PCs) devido às condições de armazenamento que favorecem a proliferação bacteriana. Neste estudo, desenvolvemos um protocolo de PCR em tempo real rápido, sensível e específico para a triagem bacteriana de PCs. Um método baseado em PCR em tempo real, controlado internamente, foi otimizado e validado com um Master Mix Universal PCR 16S (IBMP / Fiocruz), que detecta uma região conservada do gene 16S rRNA bacteriano. O background de DNA não específico foi completamente eliminado tratando a PCR Master Mix com monoazida de etídio (EMA). O limite de detecção inferior observado foi de 10 cópias equivalentes do genoma com um valor de Ct 34 ± 1,07, a curva de calibração foi gerada com diluições seriada de 10 vezes do DNA de E. coli. O tempo de processamento, incluindo a purificação microbiana do DNA, foi de aproximadamente 4 horas. O método desenvolvido mostrou alta sensibilidade sem amplificação inespecífica e menor tempo de detecção do que os métodos microbiológicos tradicionais, demonstrando ser um meio eficiente de triagem de PCs pré-transfusionais.
Palavras-chave:
contaminação bacteriana; PCR em tempo real; teste molecular; concentrado de plaquetas
1. Introduction
Bacterial contamination of blood components is one of the major causes of transfusion-related infection. Despite the implementation of preventive measures, the risk of transfusion-transmitted bacterial infection is still greater than that of transfusion-transmitted viral infection (Brecher et al., 2003BRECHER, M.E., HAY, S.N. and ROTHENBERG, S.J., 2003. Monitoring of apheresis platelet bacterial contamination with an automated liquid culture system: a university experience. Transfusion, vol. 43, no. 7, pp. 974-978. http://dx.doi.org/10.1046/j.1537-2995.2003.00438.x. PMid:12823759.
http://dx.doi.org/10.1046/j.1537-2995.20...
). The risk of receiving bacterial-contaminated platelets has been estimated to be 10 to 1,000 times higher than that of receiving platelets contaminated with viruses, such as HIV, HBV, HCV and HTLV, primarily due to the efficient screening methods used to detect viral pathogens (Blajchman, 2002BLAJCHMAN, M.A., 2002. Incidence and significance of the bacterial contamination of blood components. Developmental Biology, vol. 108, pp. 59-67. PMid:12220143.). In the United States, the residual risk of bacterial contamination is estimated at 1/6,000 for contaminated platelet products and 1/100,000 for septic reactions (Walther-Wenke, 2008WALTHER-WENKE, G., 2008. Incidence of bacterial transmission and transfusion reactions by blood components. Clinical Chemistry and Laboratory Medicine, vol. 46, no. 7, pp. 919-925. http://dx.doi.org/10.1515/CCLM.2008.151. PMid:18605950.
http://dx.doi.org/10.1515/CCLM.2008.151...
).
Transfusion therapies of blood products that are contaminated with bacteria are considered the third most common cause of death reported to the US Food and Drug Administration (FDA), following acute pulmonary lesions and hemolytic reactions related to transfusion (Razjou et al., 2017RAZJOU, F., NAGHADEH, H.T., FERDOWSI, S. and DABIRMOGHADAM, A., 2017. Evaluation of the sensitivity and specificity of use of glucose and pH for bacterial screening of platelet concentrates compared to the Bact/Alert. Indian Journal of Hematology & Blood Transfusion : An Official Journal of Indian Society of Hematology and Blood Transfusion, vol. 33, no. 1, pp. 116-120. http://dx.doi.org/10.1007/s12288-016-0660-4. PMid:28194067.
http://dx.doi.org/10.1007/s12288-016-066...
). In addition, transfusions of PCs that are contaminated with some type of bacteria can cause serious septic complications to patients.
The primary sources of such contamination include the bacterial skin flora present at puncture regions, asymptomatic bacteremic donors and contamination that occurs during product processing (Palavecino and Yomtovian, 2003PALAVECINO, E. and YOMTOVIAN, R., 2003. Risk and prevention of transfusion-related sepsis. Current Opinion in Hematology, vol. 10, no. 6, pp. 434-439. http://dx.doi.org/10.1097/00062752-200311000-00007. PMid:14564174.
http://dx.doi.org/10.1097/00062752-20031...
; Schrezenmeier et al., 2007SCHREZENMEIER, H., WALTHER-WENKE, G., MULLER, T.H., WEINAUER, F., YOUNIS, A., HOLLAND-LETZ, T., GEIS, G., ASMUS, J., BAUERFEIND, U., BURKHART, J., DEITENBECK, R., FÖRSTEMANN, E., GEBAUER, W., HÖCHSMANN, B., KARAKASSOPOULOS, A., LIEBSCHER, U.M., SÄNGER, W., SCHMIDT, M., SCHUNTER, F., SIREIS, W. and SEIFRIED, E., 2007. Bacterial contamination of platelet concentrates: results of a prospective multicenter study comparing pooled whole blood-derived platelets and apheresis platelets. Transfusion, vol. 47, no. 4, pp. 644-652. http://dx.doi.org/10.1111/j.1537-2995.2007.01166.x. PMid:17381623.
http://dx.doi.org/10.1111/j.1537-2995.20...
). The most frequent bacterial contaminants in PCs are Staphylococcus spp., Streptococcus spp., Escherichia coli, Bacillus spp., Serratia spp., Enterobacter spp., and other organisms (Wagner, 2004WAGNER, S.J., 2004. Transfusion-transmitted bacterial infection: Risks, sources and interventions. Vox Sanguinis, vol. 86, no. 3, pp. 157-163. http://dx.doi.org/10.1111/j.0042-9007.2004.00410.x. PMid:15078249.
http://dx.doi.org/10.1111/j.0042-9007.20...
).
The prevention or reduction of adverse septic reactions associated with platelets is a major challenge. There have been many advances in technologies and transfusion strategies to reduce the risk of bacterial contamination and sepsis. Some methods can efficiently identify microbial contaminants in PCs, but the time required is an inconvenience, and these methods have low specificity and sensitivity when the initial levels of bacterial contamination are low (Mancini et al., 2010MANCINI, N., CARLETTI, S., GHIDOLI, N., CICHERO, P., BURIONI, R. and CLEMENTI, M., 2010. The era of molecular and other non-culture based methods in diagnosis of sepsis. Clinical Microbiology Reviews, vol. 23, no. 1, pp. 235-251. http://dx.doi.org/10.1128/CMR.00043-09. PMid:20065332.
http://dx.doi.org/10.1128/CMR.00043-09...
; Ezuki et al., 2007EZUKI, S., KAWABATA, K., KANNO, T. and OHTO, H., 2007. Culture-based bacterial detection systems for platelets: the effect of time prior to sampling and duration of incubation required for detection with aerobic culture. Transfusion, vol. 47, no. 11, pp. 2044-2049. http://dx.doi.org/10.1111/j.1537-2995.2007.01428.x. PMid:17958533.
http://dx.doi.org/10.1111/j.1537-2995.20...
).
Culture methods are capable of detecting as few as one bacterial colony-forming unit (CFU) in a sample and can reliably detect 10 CFU in an inoculated sample without any inhibitors. In general, the BacT/ALERT system is used in Brazil for bacterial screening of PCs, which can detect 1-10 CFU per 5 mL within 24 - 48 hours (Albertoni et al., 2011ALBERTONI, G., ANDRADE, S.S., ARAUJO, P.R.B., CARVALHO, F.O., GIRÃO, M.J. and BARRETO, J.A., 2011. Evaluation of two detection methods of microorganisms in platelet concentrates. Transfusion Medicine (Oxford, England), vol. 21, no. 6, pp. 408-416. http://dx.doi.org/10.1111/j.1365-3148.2011.01105.x. PMid:21895809.
http://dx.doi.org/10.1111/j.1365-3148.20...
). The sensitivity of such tests is directly proportional to the bacteria loads and the inoculated volume in a sample. It was previously reported that increasing the sample volume from 4 to 8 mL may significantly increase the detection rate of contaminating bacteria and reduce the risk of transfusion-associated infections (Bruhn et al., 2015BRUHN, R., CUSTER, B., VANDERPOOL, S., TOWNSEND, M., KAMEL, H. and TOMASULO, P., 2015. Impact of increasing sample volume from 4 ml to 8 ml on bacterial detection rates in apheresis platelets: a metaanalysis. Vox Sanguinis, vol. 108, no. 3, pp. 318-320. http://dx.doi.org/10.1111/vox.12225. PMid:25556667.
http://dx.doi.org/10.1111/vox.12225...
). Culture-based techniques are considered the gold standard for detecting bacterial contamination in PCs. However, these methods require large sample volumes and long incubation periods that do not meet all the needs and requirements for a routine assay.
For several years, nucleic acid tests (NAT) have promised to offer more sensitive and faster alternatives to methods based on bacterial growth. These technologies function by rapidly creating copies of DNA from target cells by amplifying the nucleic acid sequences to a detectable level (Mohammadi et al., 2005MOHAMMADI, T., PIETERSZ, R.N., VANDENBROUCKE-GRAULS, C.M., SAVELKOUL, P.H. and REESINK, H.W., 2005. Detection of bacteria in platelet concentrates: comparison of broad-range real-time 16S rDNA polymerase chain reaction and automated culturing. Transfusion, vol. 45, no. 5, pp. 731-736. http://dx.doi.org/10.1111/j.1537-2995.2005.04258.x. PMid:15847662.
http://dx.doi.org/10.1111/j.1537-2995.20...
; Rood et al., 2011ROOD, I.G., PETTERSSON, A., SAVELKOUL, P.H. and DE KORTE, D., 2011. Performance and suitability of polymerase chain reaction for early detection of bacteria in platelet concentrates. Transfusion, vol. 51, no. 9, pp. 2006-2011. http://dx.doi.org/10.1111/j.1537-2995.2011.03090.x. PMid:21392020.
http://dx.doi.org/10.1111/j.1537-2995.20...
).
In a clinical context, real-time PCR is one of the most promising molecular methods for diagnosing infectious diseases with high specificity and sensitivity with a rapid turnaround time using a small sample volume. The sensitivity of real-time PCR screening allows 10-100 CFU/mL to be detected in PCs, depending on the contaminating bacterial species (Esmaili et al., 2017ESMAILI, M.A., RAZJOU, F., KHOSROSHAHI, B.N., et al, 2017. Evaluations of detections methods of bacterial contamination in platelet components. International Journal of Medical Laboratory, vol. 4, no. 4, pp. 232-245.).
Numerous studies have established the 16S rRNA gene as a universal DNA amplification target in a wide range of microorganisms (Wilson et al., 1990WILSON, K.H., BLITCHINGTON, R.B. and GREENE, R.C., 1990. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. Journal of Clinical Microbiology, vol. 28, no. 9, pp. 1942-1946. http://dx.doi.org/10.1128/JCM.28.9.1942-1946.1990. PMid:2095137.
http://dx.doi.org/10.1128/JCM.28.9.1942-...
; Anderson, 1994ANDERSON, B., 1994. Broad-range polymerase chain reaction for detection and identification of bacteria. The Journal of the Florida Medical Association, vol. 81, no. 12, pp. 835-837. PMid:7532205.; Hendolin et al., 1997HENDOLIN, P.H., MARKKANEN, A., YLIKOSKI, J. and WAHLFORS, J.J., 1997. Use of multiplex PCR for simultaneous detection of four bacterial species in middle ear effusions. Journal of Clinical Microbiology, vol. 35, no. 11, pp. 2854-2858. http://dx.doi.org/10.1128/JCM.35.11.2854-2858.1997. PMid:9350746.
http://dx.doi.org/10.1128/JCM.35.11.2854...
; Klausegger et al., 1999KLAUSEGGER, A., HELL, M., BERGER, A., ZINOBER, K., BAIER, S., JONES, N., SPERL, W. and KOFLER, B., 1999. Gram type-specific broad-range PCR amplification for rapid detection of 62 pathogenic bacteria. Journal of Clinical Microbiology, vol. 37, no. 2, pp. 464-466. http://dx.doi.org/10.1128/JCM.37.2.464-466.1999. PMid:9889245.
http://dx.doi.org/10.1128/JCM.37.2.464-4...
). However, the contamination of PCR reagents with microbial DNA is a known problem, particularly when targeting conserved regions of bacterial genomes using universal primers for broad-range PCR amplification analysis (Garson et al., 2014GARSON, J.A., PATEL, P., MCDONALD, C., BALL, J., ROSENBERG, G., TETTMAR, K.I., BRAILSFORD, S.R., PITT, T. and TEDDER, R.S. 2014. Evaluation of an ethidium monoazide-enhanced 16S rDNA real-time polymerase chain reaction assay for bacterial screening of platelet concentrates and comparison with automated culture. Transfusion, vol. 54, no. 3 Pt 2, pp. 870-878. http://dx.doi.org/10.1111/trf.12256. PMid:23701338.
http://dx.doi.org/10.1111/trf.12256...
). Because PCR can amplify low amounts of DNA, co-amplification of trace amounts of contaminating DNA can occur, producing false-positive results. Several different approaches have been described to eliminate or reduce PCR reagent contamination, such as physical, chemical and enzymatic treatments (Hein et al., 2007HEIN, I., SCHNEEWEISS, W., STANEK, C. and WAGNER, M., 2007. Ethidium monoazide and propidium monoazide for elimination of unspecific DNA background in quantitative universal real-time PCR. Journal of Microbiological Methods, vol. 71, no. 3, pp. 336-339. http://dx.doi.org/10.1016/j.mimet.2007.09.005. PMid:17936386.
http://dx.doi.org/10.1016/j.mimet.2007.0...
; Humphrey et al., 2015HUMPHREY, B., MCLEOD, N., TURNER, C., SUTTON, J.M., DARK, P.M. and WARHURST, G., 2015. Removal of contaminant DNA by combined UV-EMA treatment allows low copy number detection of clinically relevant bacteria using pan-bacterial real-time PCR. PLoS One, vol. 10, no. 7, pp. e0132954. http://dx.doi.org/10.1371/journal.pone.0132954. PMid:26172943.
http://dx.doi.org/10.1371/journal.pone.0...
). The treatment of PCR master mixes with ethidium monoazide (EMA) followed by photoactivation is considered to be the most reliable and effective means of eliminating residual contaminating DNA without compromising the sensitivity of the assay (Hein et al., 2007HEIN, I., SCHNEEWEISS, W., STANEK, C. and WAGNER, M., 2007. Ethidium monoazide and propidium monoazide for elimination of unspecific DNA background in quantitative universal real-time PCR. Journal of Microbiological Methods, vol. 71, no. 3, pp. 336-339. http://dx.doi.org/10.1016/j.mimet.2007.09.005. PMid:17936386.
http://dx.doi.org/10.1016/j.mimet.2007.0...
; Humphrey et al., 2015HUMPHREY, B., MCLEOD, N., TURNER, C., SUTTON, J.M., DARK, P.M. and WARHURST, G., 2015. Removal of contaminant DNA by combined UV-EMA treatment allows low copy number detection of clinically relevant bacteria using pan-bacterial real-time PCR. PLoS One, vol. 10, no. 7, pp. e0132954. http://dx.doi.org/10.1371/journal.pone.0132954. PMid:26172943.
http://dx.doi.org/10.1371/journal.pone.0...
; Rueckert and Morgan, 2007RUECKERT, A. and MORGAN, H.W., 2007. Removal contaminating DNA from polymerase chain reaction using ethidium monoazide. Journal of Microbiological Methods, vol. 68, no. 3, pp. 596-600. http://dx.doi.org/10.1016/j.mimet.2006.11.006. PMid:17187883.
http://dx.doi.org/10.1016/j.mimet.2006.1...
; Patel et al., 2012PATEL, P., GARSON, J.A., TETTMAR, K.I., ANCLIFF, S., MCDONALD, C., PITT, T., COELHO, J. and TEDDER, R.S., 2012. Development of an ethidium monoazide-enhanced internally controlled universal 16S rDNA real-time polymerase chain reaction assay for detection of bacterial contamination in platelet concentrates. Transfusion, vol. 52, no. 7, pp. 1423-1432. http://dx.doi.org/10.1111/j.1537-2995.2011.03484.x. PMid:22188457.
http://dx.doi.org/10.1111/j.1537-2995.20...
; Takahashi et al., 2014TAKAHASHI, H., YAMAZAKI, H., AKANUMA, S., KANAHARA, H., SAITO, T., CHIMURO, T., KOBAYASHI, T., OHTANI, T., YAMAMOTO, K., SUGIYAMA, S. and KOBORI, T., 2014. Preparation of Phi29 DNA Polymerase free of Amplifiable DNA using Ethidium Monoazide, an ultraviolet-free light-emitting diode lamp and trehalose. PLoS One, vol. 9, no. 2, pp. e82624. http://dx.doi.org/10.1371/journal.pone.0082624. PMid:24505243.
http://dx.doi.org/10.1371/journal.pone.0...
).
In this study, we developed a simple, rapid and sensitive broad-range real-time PCR protocol for bacterial screening of PCs that eliminates the problem of co-amplification of contaminating microbial DNA. This protocol will be particularly useful for assaying samples with low levels of contamination and to detect microorganisms that are difficult to grow in vitro or require a long period of incubation.
2. Materials and Methods
2.1. Real-Time PCR
2.1.1. Design and optimization
A broad-range bacterial PCR detection system targeting a highly conserved region of the 16S rRNA gene was previously described (Yang et al., 2002YANG, S., LIN, S., KELEN, G.D., QUINN, T.C., DICK, J.D., GAYDOS, C.A. and ROTHMAN, R.E., 2002. Quantitative multiprobe PCR assay for simultaneous detection and identification to species level of bacterial pathogens. Journal of Clinical Microbiology, vol. 40, no. 9, pp. 3449-3454. http://dx.doi.org/10.1128/JCM.40.9.3449-3454.2002. PMid:12202592.
http://dx.doi.org/10.1128/JCM.40.9.3449-...
). Representative 16S rRNA gene sequences for the most common bacterial species causing platelet concentrate contamination were obtained from the GenBank sequence database (www.ncbi.nlm.nih.gov/genbank/ ) (including Propionibacterium acnes, Bacillus cereus, Staphylococcus aureus, Staphylococcus epidermidis, Escherichia coli, Serratia marcescens and Klebsiella pneumoniae). The sequences were aligned using MEGA 7 (Kumar et al., 2016KUMAR, S., STECHER, G. and TAMURA, K., 2016. MEGA 7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, vol. 33, no. 7, pp. 1870-1874. http://dx.doi.org/10.1093/molbev/msw054. PMid:27004904.
http://dx.doi.org/10.1093/molbev/msw054...
). The forward primer sequence was modified to allow Propionibacterium acnes to be detected. The optimized oligonucleotide concentrations used were 500 nM of each PCR primer and 250 nM of the hydrolysis probe. The human ribonuclease P gene (encoding RNase P) amplification system was included as internal positive control using 800 nM of each PCR primer and 50 nM of the hydrolysis probe according to a previously described protocol (WHO, 2009WORLD HEALTH ORGANIZATION – WHO, 2009. CDC protocol of real time RTPCR for swine influenza A(H1N1). Geneva: WHO, pp. 1-9.). Reactions were performed in a 20 µL final volume using the commercial TaqMan Universal PCR Master Mix (Thermo Fisher Scientific) containing DNA purified from microbial pure culture. A minimum of three reactions were performed for each experiment using the following cycling condition: 95°C for 10 min followed by 40 cycles of 95 °C for 15 sec and 60 °C for 1 min.
2.1.2 Proprietary reaction composition
A proprietary reaction mixture was optimized from a basic PCR reaction buffer (10 mM Tris-HCl (pH 8.8), 50 mM KCl, 0.1% Triton X-100, and 0.1 mM DTT), supplemented with 1.5 mM MgSO4, 0.2 mM dNTPs, 30 nM ROX and 2 units of hot start Taq DNA polymerase. Other salts (MgCl2 or MgAc) and their concentrations (from 1.5 to 9 mM), as well as additives (BSA, ammonium acetate, potassium glutamate, potassium sulfate and betaine) were also evaluated but did not improve the detection limits or reaction efficiencies. A commercial TaqMan Universal PCR Master Mix (Thermo Fisher Scientific) was used as a reference. The final optimized real-time PCR reaction mixture was produced in our GMP (good manufacturing practices) facility according to quality standards for diagnostics applied to health. Oligonucleotides were synthesized by Integrated DNA Technologies, Inc. (IDT) and were purified by HPLC.
2.1.3. Analytical performance
DNA was purified from pure microbial cultures and used to determine the analytical performance of the bacterial 16S rRNA gene detection assay. The purity and quantity of each DNA sample was measured using a DeNovix DS-11+ spectrophotometer (DeNovix Inc., Wilmington, DE, USA). The quantified DNA was diluted 10-fold (107 to 101 genome equivalents per 5 µL) and 5 µL of each dilution was used as template DNA in PCR reactions to generate the calibration curve. As a control, 103 genome equivalents of human DNA was assayed.
2.1.4. Removal of contaminant DNA
PCR Master Mix reactions were treated with ethidium monoazide (EMA) as described (Patel et al., 2012PATEL, P., GARSON, J.A., TETTMAR, K.I., ANCLIFF, S., MCDONALD, C., PITT, T., COELHO, J. and TEDDER, R.S., 2012. Development of an ethidium monoazide-enhanced internally controlled universal 16S rDNA real-time polymerase chain reaction assay for detection of bacterial contamination in platelet concentrates. Transfusion, vol. 52, no. 7, pp. 1423-1432. http://dx.doi.org/10.1111/j.1537-2995.2011.03484.x. PMid:22188457.
http://dx.doi.org/10.1111/j.1537-2995.20...
) to overcome the contamination of PCR reagents with residual microbial DNA. Briefly, EMA (Sigma-Aldrich) was dissolved in 20% DMSO at a concentration of 10 mg/mL in the dark, after which 50-µL aliquots were stored at -20 °C. Working solutions were prepared fresh on the day of use by dilution in molecular biology grade water (Bioline). The complete PCR Master Mix, including primers and probes, were treated with 1.2 µM EMA and photoactivated on ice with a 500 W halogen light source (Golden) for 5 min at a distance of approximately 20 cm from the tubes. Next, the PCR Master Mix was distributed in the 96-well plate (15 µL), wells were loaded with 5 µL of DNA template or water (no template control, NTC) and real-time PCR was performed on a 7500 Real Time PCR System (Thermo Fisher Scientific). Assays were performed using a minimum of three experimental replicates. Statistical analyses (standard curve, mean ± standard deviation, 95% confidence intervals (CI), coefficient of variation) were calculated using GraphPad Prism v6.0 (Graph Pad Prism Inc., USA).
2.2. DNA extraction
Platelet concentrates were processed targeting the enrichment of microbial DNA using a selective lysis approach followed by the disruption of microbial cells with an alkaline solution as described previously (Dobbelaer et al., 2012DOBBELAER, I., NEERKEN, S., VAN DE WIEL, P., VAN MEERBERGEN, B., and PENTERMAN, R. Selective lysis of cells by ionic surfactants. nº WO2012168003 A1. 13-12-2012.; Van Meerbergen et al., 2011VAN MEERBERGEN, B.E.G.J., PICIU, O.M., GIL, R., SCHMIDT, K.A., NEERKEN, S., PONJEE, M.W.G., UNAY, Z.S., PENTERMAN, R. and VAN DE WIEL, P.A. Selective lysis of cell. nº WO2011070507 A1. 16-06-2011.; Loonen et al., 2013LOONEN, A. J. M., BOS, M. P., VAN MEERBERGEN, B., NEERKEN, S., CATSBURG, A., DOBBELAER, I., PENTERMAN, R., MAERTENS, G., VAN DE WIEL, P., SAVELKOUL, P., and VAN DEN BRULE, A. J. C., 2013. Comparison of pathogen DNA isolation methods from large volumes of whole blood to improve molecular diagnosis of bloodstream infections. Plos One, vol. 8, no. 8, pp. e72349.; Trung et al., 2016TRUNG, N.T., HIEN, T.T.T., HUYEN, T.T.T., QUYEN, D.T., VAN SON, T., HOAN, P.Q., PHUONG, N.T., LIEN, T.T., BINH, M.T., VAN TONG, H., MEYER, C.G., VELAVAN, T.P. and SONG, H., 2016. Enrichment of bacterial DNA for the diagnosis of blood stream infections. BMC Infectious Diseases, vol. 16, no. 1, pp. 235. http://dx.doi.org/10.1186/s12879-016-1568-1. PMid:27246723.
http://dx.doi.org/10.1186/s12879-016-156...
). Five milliliters of pooled whole blood-derived PCs (1 mL from each of 5 individual donors) was mixed with an equal volume of selective lysis buffer (500 mM sodium carbonate, 1% Triton X-100, pH 10.5) by inverting the tubes for 30 seconds. The selective lysis step was stopped by the addition of an equal volume of initial sample volume of neutralization buffer (1 M Tris). Intact microbial cells were then concentrated by centrifuging the suspension for 15 minutes at 2,791 xg. Pellets were resuspended in 5 mL of washing buffer (1X phosphate buffered saline - PBS) and then centrifuged for 15 minutes at maximum speed. The resulting pellets were then resuspended in 200 µL of alkaline lysis buffer (200 mM NaOH and 0.5% SDS) and incubated for 10 minutes at 95 °C in a thermomixer set at 1,000 rpm. Finally, 20 µL of neutralization buffer (1 M Citric Acid Solution) was added and the microbial DNA was purified using a QIAamp Blood Mini kit (QIAGEN) according to the manufacturer’s protocol.
3. Results
3.1. Primers and probe design
We evaluated primers and probes described in literature that were primarily developed for universal/pan-bacterial PCR detection. A bioinformatics analysis was performed by aligning 16S rRNA gene sequences of the major bacteria that contaminate platelet concentrates and cause bloodstream infections. A set of universal oligonucleotides specific for the conserved 16S rRNA gene is displayed in Figure 1, showing the P891F-modified primer designed in this study to incorporate the P. acnes target sequence.
Representative nucleotide sequence alignment of the bacterial 16S rRNA gene PCR target. Primers and probes are depicted in the picture. Propionibacterium acnes (accession number AB108484), Klebsiella pneumoniae (accession number NC_009648), Serratia marcescens (accession number NC_005211), Escherichia coli (accession number AE005174), Bacillus cereus (accession number AP007209), Staphylococcus aureus (accession number AP017922) and Staphylococcus epidermidis (accession number NC_002976). Sequences were obtained from the GenBank database.
For primers and probe evaluations, PCR assays were performed with commercial TaqMan Universal PCR Master Mix (Thermo Fisher Scientific). Amplification efficiencies obtained with the forward reference primer (P891F) were compared to the newly designed primer (P891F-modified) using DNA that was purified from pure cultures of P. acnes or E. coli. The results show that the P891F-modified primer is slightly more efficient than P891F for P. acnes DNA amplification (Figure 2A). No difference between the two primers was observed when E. coli DNA was used as a template (Figure 2B).
Comparison of bacterial 16S rRNA gene amplification efficiencies using the primers P891F (black line) and P891F-modified (gray line). The results were obtained using 10-fold serial dilutions containing 106 to 103 genome equivalents per reaction. (A) P. acnes DNA. (B) E. coli DNA.
3.2. Real-time PCR optimization
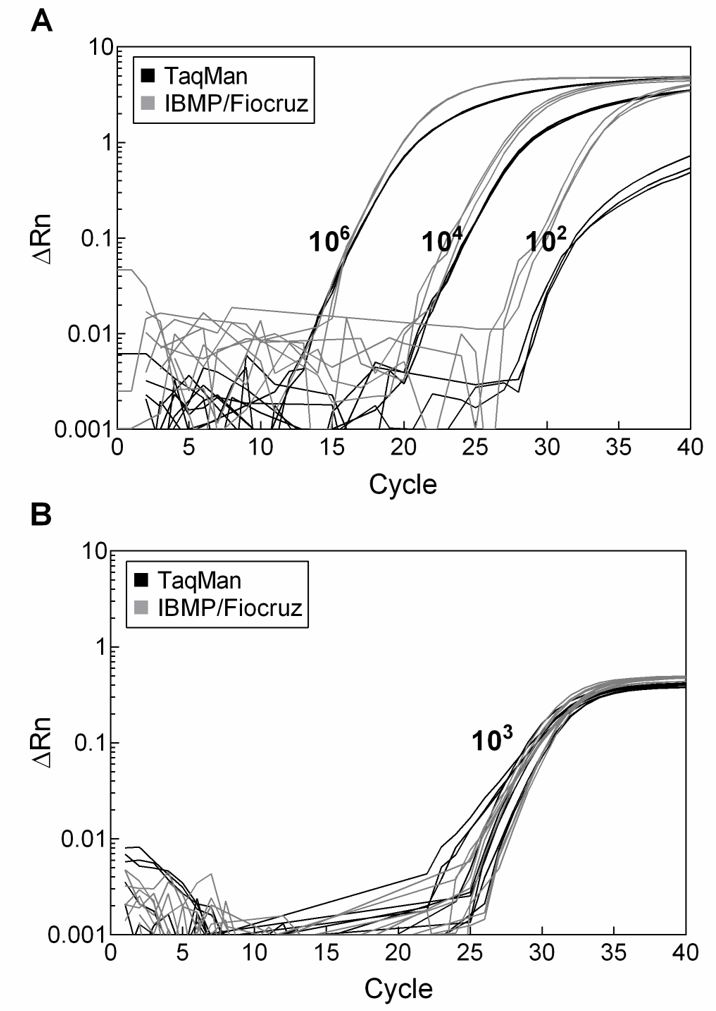
We evaluated the effect of several PCR cofactors on the amplification efficiencies of the bacterial 16S rRNA gene in the presence of human genomic DNA. The best experimental optimized condition was named 16S Universal PCR Master Mix (IBMP/Fiocruz) and was compared to the TaqMan Universal PCR Master Mix (Thermo Fisher Scientific) for PCR amplification efficiency. The results show a more efficient reaction with the optimized IBMP/Fiocruz proprietary Master Mix formulation, especially with low quantities of target DNA (Figure 3). As the target quantity decreased, the reaction curve generated with our 16S Universal PCR Master Mix shift to the left compared to the commercial TaqMan Universal PCR Master Mix, presenting lower Ct values (Figure 3A). The human RNase P-encoding gene was amplified in each reaction as an internal control (Figure 3B).
Comparison of bacterial 16S gene amplification with 16S Universal PCR Master Mix - IBMP/Fiocruz (grey line) and TaqMan Universal PCR Master Mix - Thermo Fisher Scientific (black line). (A) Amplification plot obtained with a known concentration of E. coli DNA (106, 104 and 102 genome equivalents per reaction) in the presence of 103 copies of human genomic DNA per reaction. (B) Amplification of the human RNase P-encoding gene.
3.3. Analytical sensitivity test
Using a 10-fold dilution calibration curve covering a range of 107 to 101 genome equivalents per reaction, a linear dynamic range was established with a strong linear correlation (R2 = 0.999) (Figure 4A). The limit of detection (LoD) of the bacterial 16S rRNA gene, concomitant with the amplification of the human RNase P-encoding gene in duplex reactions, was calculated from the lower concentration with a 95% rate of detection. The LoD was set at 10 bacterial genome equivalents with a Ct value of 34±1.07. GraphPad Prism was set the cut-off value with two standard deviations above the lower LoD detected (95% CI), so the Ct values > 36.19 indicating a negative result.
Calibration curve generated with 10-fold serial dilutions of E. coli DNA. (A) The calibration curve containing 107 to 101 genome equivalents per reaction. The assay showed a strong linear dynamic range over 6 log10 concentrations with R2 = 0.999. The mean reaction efficiency was 90% with a slope of -3.584 and a y-intercept at 37.24. Dashed line represents the cut-off value (Ct 36.10). The graph was generated using six experimental replicates. (B) Comparison of linear regression of the threshold cycles (Ct) to detect the E. coli 16S rRNA gene with PCR Master Mix that was untreated (black square) or treated (grey circle) with ethidium monoazide (EMA). The calibration curve was generated using 10-fold serial dilutions containing 105 to 101 genome equivalents per reaction. Paired t-test: P > 0.05 (0.2909).
3.4. PCR Master Mix decontamination via EMA treatment
First, the analytical sensitivity of the assay untreated or treated with EMA was evaluated with a 10-fold serial dilution of bacterial DNA from 105 to 101 genome equivalents per reaction. The untreated and treated with 1.2 μM EMA show equivalent amplification profiles as observed in Figure 4B. No significant difference between the conditions was observed when analyzing the amplification yield or sensitivity of the assay.
Next, the 16S Universal PCR Master Mix that was untreated or treated with 1.2 μM EMA was evaluated for non-specific amplification in no template control reactions (NTC). Each condition was analyzed using 48 replicates in the same 96-well plate. As show in the Figure 5, no amplification was observed using the Master Mix treated with EMA, while the untreated Master Mix presented non-specific amplification signals with a minimum Ct of 35.45. The results showed a significant difference between untreated and treated Master Mix conditions (p = 0.0001) (Fisher, 1956FISHER, R.A., 1956. Statistical methods for research workers. 12th ed. London: Quarterly Journal of the Royal Meteorol Society.).
Evaluation of non-specific bacterial 16S rRNA gene amplification in no template control (NTC) reactions using PCR Master Mix that was untreated or treated with ethidium monoazide (EMA).
3.5. Routine platelet screening
This study was performed with PCs from HEMEPAR (Center of Hematology and Haemotherapy of Paraná, Curitiba, Brazil) and was approved by the Institutional Review Board of Hospital do Trabalhador/SES/PR (IRB# 51711815.0.0000.5225). The 16S PCR-based method to screen platelet concentrates for bacteria was evaluated in routine samples undertaken at 24 hours after donation. A total of 250 PCs that were considered negative for bacterial contamination, according to standard protocol applied in the HEMEPAR routine, were analyzed in pools containing five PCs each sample, resulting in 50 samples. The samples were processed on 12 consecutive working days, the results of which are show in Figure 6. A Ct distribution above 37 was observed for the 50 samples assayed, considering that all samples were negative for bacterial contamination. For each assay, no template control (NTC) did not show any amplification and positive control with pre-determined bacterial concentration reactions were evaluated.
Evaluation of bacterial 16S rRNA gene detection in routine platelet concentrates from HEMEPAR. The total number corresponds to 50 samples analyzed on 12 consecutive working days. NTC amplification is shown for each of the 12 days of analysis. The dashed line represents the cut-off value (Ct 36.10).
4. Discussion
Several techniques have been adopted to prevent or reduce transfusion-related bloodstream infections associated with blood banks, varying from manual methods to automated culture systems. These methods have been demonstrated to be useful for bacterial detection in platelet concentrates, but these systems can fail to produce timely results due to the time required to indicate bacteria presence (Macauley et al., 2003MACAULEY, A., CHANDRASEKAR, A., GEDDIS, G., MORRIS, K.G. and MCCLELLAND, W.M., 2003. Operational feasibility of routine bacterial monitoring of platelets. Transfusion Medicine (Oxford, England), vol. 13, no. 4, pp. 189-195. http://dx.doi.org/10.1046/j.1365-3148.2003.00441.x. PMid:12880389.
http://dx.doi.org/10.1046/j.1365-3148.20...
; Ecker et al., 2010ECKER, D.J., SAMPATH, R., LI, H., MASSIRE, C., MATTHEWS, H.E., TOLENO, D., HALL, T.A., BLYN, L.B., ESHOO, M.W., RANKEN, R., HOFSTADLER, S.A. and TANG, Y.W., 2010. New technology for rapid molecular diagnosis of bloodstream infections. Expert Review of Molecular Diagnostics, vol. 10, no. 4, pp. 399-415. http://dx.doi.org/10.1586/erm.10.24. PMid:20465496.
http://dx.doi.org/10.1586/erm.10.24...
; Pietersz et al., 2003PIETERSZ, R.N., ENGELFRIET, C.P. and REESINK, H.W., 2003. Detection of bacterial contamination of platelet concentrates. Vox Sanguinis, vol. 85, no. 3, pp. 224-239. PMid:14516456.).
Culture methods continue to be the primary means for bacterial screening, and commercially available automated systems are the most commonly used method for the quality control of blood components. However, in many cases, the patient may be transfused before the culture results are obtained from automated culture systems. Samples containing slow-growing organisms and low bacterial loads are particularly prone to producing false-negative culture results (Benjamin and Wagner, 2007BENJAMIN, R.J. and WAGNER, S.J., 2007. The residual risk of sepsis: modelling the effect of concentration on bacterial detection in two-bottle culture systems and an estimation of false negative culture rates. Transfusion, vol. 47, no. 8, pp. 1381-1389. http://dx.doi.org/10.1111/j.1537-2995.2007.01326.x. PMid:17655581.
http://dx.doi.org/10.1111/j.1537-2995.20...
). In addition, culture methods miss fastidious organisms that are difficult or impossible to culture.
Therefore, new strategies have been proposed to reduce the risk of transfusion-associated bloodstream infections (Blajchman et al., 2004BLAJCHMAN, M.A., GOLDMAN, M. and BAEZA, F., 2004. Improving the bacteriological safety of platelet transfusions. Transfusion Medicine Reviews, vol. 18, no. 1, pp. 11-24. http://dx.doi.org/10.1016/j.tmrv.2003.10.002. PMid:14689374.
http://dx.doi.org/10.1016/j.tmrv.2003.10...
). Advances have been made in molecular biology that offer more sensitive and fast alternatives to traditional methods, and PCR-based methods are considered the most promising among them (Paolucci et al., 2010PAOLUCCI, M., LANDINI, M.P. and SAMBRI, V., 2010. Conventional and molecular techniques for the early diagnosis of bacteraemia. International Journal of Antimicrobial Agents, vol. 36, suppl. 2, pp. S6-S16. http://dx.doi.org/10.1016/j.ijantimicag.2010.11.010. PMid:21129933.
http://dx.doi.org/10.1016/j.ijantimicag....
). According to the literature, a real-time PCR should be simple, sensitive enough to detect clinically significant levels of bacteria, highly specific, and rapid enough to allow platelet concentrates to be cleared for clinical use within hours (Mohammadi et al., 2006MOHAMMADI, T., SAVELKOUL, P.H., PIETERSZ, R.N. and REESINK, H.W., 2006. Applications of real-time PCR in the screening of platelet concentrates for bacterial contamination. Expert Review of Molecular Diagnostics, vol. 6, no. 6, pp. 865-872. http://dx.doi.org/10.1586/14737159.6.6.865. PMid:17140373.
http://dx.doi.org/10.1586/14737159.6.6.8...
; Corless et al., 2000CORLESS, C.E., GUIVER, M., BORROW, R., EDWARDS-JONES, V., KACZMARSKI, E.B. and FOX, A.J., 2000. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. Journal of Clinical Microbiology, vol. 38, no. 5, pp. 47-52. http://dx.doi.org/10.1128/JCM.38.5.1747-1752.2000. PMid:10790092.
http://dx.doi.org/10.1128/JCM.38.5.1747-...
). Furthermore, real-time PCR allows the number of CFU present in a sample to be estimated by interpolating the Ct value in a linear regression formula that describes a dynamic range of the quantifiable copies established by a calibration curve (Bustin et al., 2009BUSTIN, S.A., BENES, V., GARSON, J.A., HELLEMANS, J., HUGGETT, J., KUBISTA, M., MUELLER, R., NOLAN, T., PFAFFL, M.W., SHIPLEY, G.L., VANDESOMPELE, J. and WITTWER, C.T., 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry, vol. 55, no. 4, pp. 611-622. http://dx.doi.org/10.1373/clinchem.2008.112797. PMid:19246619.
http://dx.doi.org/10.1373/clinchem.2008....
).
In this study, we modified the primer-probe set originally described by Yang and coworkers (Yang et al., 2002YANG, S., LIN, S., KELEN, G.D., QUINN, T.C., DICK, J.D., GAYDOS, C.A. and ROTHMAN, R.E., 2002. Quantitative multiprobe PCR assay for simultaneous detection and identification to species level of bacterial pathogens. Journal of Clinical Microbiology, vol. 40, no. 9, pp. 3449-3454. http://dx.doi.org/10.1128/JCM.40.9.3449-3454.2002. PMid:12202592.
http://dx.doi.org/10.1128/JCM.40.9.3449-...
). The forward primer was adapted to allow for the detection of Propionibacterium acnes, one of the most common contaminating microbial species in PCs and which was previously described using an additional set of oligonucleotides specific for its detection via PCR reaction (Patel et al., 2012PATEL, P., GARSON, J.A., TETTMAR, K.I., ANCLIFF, S., MCDONALD, C., PITT, T., COELHO, J. and TEDDER, R.S., 2012. Development of an ethidium monoazide-enhanced internally controlled universal 16S rDNA real-time polymerase chain reaction assay for detection of bacterial contamination in platelet concentrates. Transfusion, vol. 52, no. 7, pp. 1423-1432. http://dx.doi.org/10.1111/j.1537-2995.2011.03484.x. PMid:22188457.
http://dx.doi.org/10.1111/j.1537-2995.20...
). Moreover, we developed a test with a proprietary reaction mixture that exhibits better efficiency and sensitivity than the commercial TaqMan Universal Master Mix (Thermo Fisher Scientific).
According to earlier studies, virtually all PCR reagents are contaminated with minute amounts of bacterial DNA (Corless et al., 2000CORLESS, C.E., GUIVER, M., BORROW, R., EDWARDS-JONES, V., KACZMARSKI, E.B. and FOX, A.J., 2000. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. Journal of Clinical Microbiology, vol. 38, no. 5, pp. 47-52. http://dx.doi.org/10.1128/JCM.38.5.1747-1752.2000. PMid:10790092.
http://dx.doi.org/10.1128/JCM.38.5.1747-...
). This finding is problematic for highly sensitive PCR-based assays, especially when detecting highly conserved regions in the bacterial genome, such as the 16S rRNA gene (Albertoni et al., 2011ALBERTONI, G., ANDRADE, S.S., ARAUJO, P.R.B., CARVALHO, F.O., GIRÃO, M.J. and BARRETO, J.A., 2011. Evaluation of two detection methods of microorganisms in platelet concentrates. Transfusion Medicine (Oxford, England), vol. 21, no. 6, pp. 408-416. http://dx.doi.org/10.1111/j.1365-3148.2011.01105.x. PMid:21895809.
http://dx.doi.org/10.1111/j.1365-3148.20...
). Because the frequency of bacterial contamination of platelet concentrates is low and the product is scarce, it is necessary to minimize false-positive results. In this context, the primary parameters that need to be addressed are the analytical sensitivity and specificity of the assay, which refers to how many target copies the assay is able to detect and whether the no template controls (NTCs) are reliably negative. Determining the sensitivity and specificity equilibrium is a fundamental of evaluating a prototype test for use in blood centers. The treatment of PCR reagents with EMA is a reliable means of reducing the level of residual DNA contamination in these products, which we confirmed to be an effective solution in this study.
To overcome the difficulties associated with the limit of detection calculation described in the literature, we considered the analytical sensitivity of the test as the lowest copy number detected in 95% of the reactions (Bustin et al., 2009BUSTIN, S.A., BENES, V., GARSON, J.A., HELLEMANS, J., HUGGETT, J., KUBISTA, M., MUELLER, R., NOLAN, T., PFAFFL, M.W., SHIPLEY, G.L., VANDESOMPELE, J. and WITTWER, C.T., 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry, vol. 55, no. 4, pp. 611-622. http://dx.doi.org/10.1373/clinchem.2008.112797. PMid:19246619.
http://dx.doi.org/10.1373/clinchem.2008....
), which can also be interpreted as the lowest copy number that can be distinguished from background (Armbruster et al., 1994ARMBRUSTER, D.A., TILLMAN, M.D. and HUBBS, L.M., 1994. Limit of detection (LOD)/limit of quantification (LOQ): comparison of the empirical and the statistical methods exemplified with GC-MS assays of abused drugs. Clinical Chemistry, vol. 40, no. 7, pp. 1233-1238. http://dx.doi.org/10.1093/clinchem/40.7.1233. PMid:8013092.
http://dx.doi.org/10.1093/clinchem/40.7....
; Burns and Valdivia, 2008BURNS, M. and VALDIVIA, H., 2008. Modelling the limit of detection in real-time quantitative PCR. European Food Research and Technology, vol. 226, no. 6, pp. 1513-1524. http://dx.doi.org/10.1007/s00217-007-0683-z.
http://dx.doi.org/10.1007/s00217-007-068...
). The limit of detection estimates in real-time PCR-based methods are particularly complicated when the template concentration is zero, because Ct is undefined and determination of a LoD algorithm is the focus of continuous research (Burns and Valdivia, 2008BURNS, M. and VALDIVIA, H., 2008. Modelling the limit of detection in real-time quantitative PCR. European Food Research and Technology, vol. 226, no. 6, pp. 1513-1524. http://dx.doi.org/10.1007/s00217-007-0683-z.
http://dx.doi.org/10.1007/s00217-007-068...
). Considering the PCR approach, the most sensitive LoD that is theoretically possible is 3 copies of a template per reaction (Bustin et al., 2009BUSTIN, S.A., BENES, V., GARSON, J.A., HELLEMANS, J., HUGGETT, J., KUBISTA, M., MUELLER, R., NOLAN, T., PFAFFL, M.W., SHIPLEY, G.L., VANDESOMPELE, J. and WITTWER, C.T., 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry, vol. 55, no. 4, pp. 611-622. http://dx.doi.org/10.1373/clinchem.2008.112797. PMid:19246619.
http://dx.doi.org/10.1373/clinchem.2008....
). We calculated a mean for each data set in the calibration curve and analyzed the lowest detectable point corresponding to 10 bacterial genome equivalents per reaction with a 100% of detection and a cut-off value established with two standard deviations above.
The platelet concentrate samples analyzed according to the established HEMEPAR protocol were considered negative and inserted into the donation routine. The 16S rRNA gene PCR-based method described was used to assay 250 routine PCs, and the results confirmed that all samples were negative for bacterial contamination, although some samples crossed the threshold before 40 cycles with a lower detection point above the established cut-off. These negative results were expected, since approximately 1 in 1,000 to 3,000 platelets units may be contaminated with bacteria (Blajchman et al., 2005BLAJCHMAN, M.A., BECKERS, E.A., DICKMEISS, E., LIN, L., MOORE, G. and MUYLLE, L., 2005. Bacterial detection of platelets: current problems and possible resolutions. Transfusion Medicine Reviews, vol. 19, no. 4, pp. 259-272. http://dx.doi.org/10.1016/j.tmrv.2005.05.002. PMid:16214015.
http://dx.doi.org/10.1016/j.tmrv.2005.05...
; Palavecino et al., 2006PALAVECINO, E.L., YOMTOVIAN, R.A. and JACOBS, M.R., 2006. Detecting bacterial contamination in platelet products. Clinical Laboratory, vol. 56, no. 9-10, pp. 443-456. PMid:17078471.; Das et al., 2015DAS, S., KALE, M., BEENA, P.M. and KUMAR, H., 2015. Bacterial contamination of platelet at University Hospital: “A prospective surveillance study. INSS, vol. 4, no. 4, pp. 505-508.).
Molecular techniques can provide information regarding blood contamination during collection or processing and can be implemented in blood banks as a routine screening test to reduce and prevent the risk of transfusion-transmitted bacterial infections. Additionally, this information can be valuable from a statistical and epidemiological point of view, as it could be an opportunity to gain knowledge regarding the contamination detection sources (Mohammadi et al., 2006MOHAMMADI, T., SAVELKOUL, P.H., PIETERSZ, R.N. and REESINK, H.W., 2006. Applications of real-time PCR in the screening of platelet concentrates for bacterial contamination. Expert Review of Molecular Diagnostics, vol. 6, no. 6, pp. 865-872. http://dx.doi.org/10.1586/14737159.6.6.865. PMid:17140373.
http://dx.doi.org/10.1586/14737159.6.6.8...
).
In conclusion, the broad-range real-time PCR-based method described in this study may be used to screen for bacteria in pre-transfusion platelet concentrates. Furthermore, real-time PCR can be integrated into automated sample processing platforms to allow for complete automation of the process, meeting the requirements for blood components screening in blood centers.
Acknowledgements
We thank Analia Maria Breckenfeld Machado and Adriana Nascimento de Araujo from HEMEPAR for providing routine platelet concentrate samples. This work was supported by the Fundação Oswaldo Cruz.
-
(With 6 figures)
References
- ALBERTONI, G., ANDRADE, S.S., ARAUJO, P.R.B., CARVALHO, F.O., GIRÃO, M.J. and BARRETO, J.A., 2011. Evaluation of two detection methods of microorganisms in platelet concentrates. Transfusion Medicine (Oxford, England), vol. 21, no. 6, pp. 408-416. http://dx.doi.org/10.1111/j.1365-3148.2011.01105.x PMid:21895809.
» http://dx.doi.org/10.1111/j.1365-3148.2011.01105.x - ANDERSON, B., 1994. Broad-range polymerase chain reaction for detection and identification of bacteria. The Journal of the Florida Medical Association, vol. 81, no. 12, pp. 835-837. PMid:7532205.
- ARMBRUSTER, D.A., TILLMAN, M.D. and HUBBS, L.M., 1994. Limit of detection (LOD)/limit of quantification (LOQ): comparison of the empirical and the statistical methods exemplified with GC-MS assays of abused drugs. Clinical Chemistry, vol. 40, no. 7, pp. 1233-1238. http://dx.doi.org/10.1093/clinchem/40.7.1233 PMid:8013092.
» http://dx.doi.org/10.1093/clinchem/40.7.1233 - BENJAMIN, R.J. and WAGNER, S.J., 2007. The residual risk of sepsis: modelling the effect of concentration on bacterial detection in two-bottle culture systems and an estimation of false negative culture rates. Transfusion, vol. 47, no. 8, pp. 1381-1389. http://dx.doi.org/10.1111/j.1537-2995.2007.01326.x PMid:17655581.
» http://dx.doi.org/10.1111/j.1537-2995.2007.01326.x - BLAJCHMAN, M.A., 2002. Incidence and significance of the bacterial contamination of blood components. Developmental Biology, vol. 108, pp. 59-67. PMid:12220143.
- BLAJCHMAN, M.A., BECKERS, E.A., DICKMEISS, E., LIN, L., MOORE, G. and MUYLLE, L., 2005. Bacterial detection of platelets: current problems and possible resolutions. Transfusion Medicine Reviews, vol. 19, no. 4, pp. 259-272. http://dx.doi.org/10.1016/j.tmrv.2005.05.002 PMid:16214015.
» http://dx.doi.org/10.1016/j.tmrv.2005.05.002 - BLAJCHMAN, M.A., GOLDMAN, M. and BAEZA, F., 2004. Improving the bacteriological safety of platelet transfusions. Transfusion Medicine Reviews, vol. 18, no. 1, pp. 11-24. http://dx.doi.org/10.1016/j.tmrv.2003.10.002 PMid:14689374.
» http://dx.doi.org/10.1016/j.tmrv.2003.10.002 - BRECHER, M.E., HAY, S.N. and ROTHENBERG, S.J., 2003. Monitoring of apheresis platelet bacterial contamination with an automated liquid culture system: a university experience. Transfusion, vol. 43, no. 7, pp. 974-978. http://dx.doi.org/10.1046/j.1537-2995.2003.00438.x PMid:12823759.
» http://dx.doi.org/10.1046/j.1537-2995.2003.00438.x - BRUHN, R., CUSTER, B., VANDERPOOL, S., TOWNSEND, M., KAMEL, H. and TOMASULO, P., 2015. Impact of increasing sample volume from 4 ml to 8 ml on bacterial detection rates in apheresis platelets: a metaanalysis. Vox Sanguinis, vol. 108, no. 3, pp. 318-320. http://dx.doi.org/10.1111/vox.12225 PMid:25556667.
» http://dx.doi.org/10.1111/vox.12225 - BURNS, M. and VALDIVIA, H., 2008. Modelling the limit of detection in real-time quantitative PCR. European Food Research and Technology, vol. 226, no. 6, pp. 1513-1524. http://dx.doi.org/10.1007/s00217-007-0683-z
» http://dx.doi.org/10.1007/s00217-007-0683-z - BUSTIN, S.A., BENES, V., GARSON, J.A., HELLEMANS, J., HUGGETT, J., KUBISTA, M., MUELLER, R., NOLAN, T., PFAFFL, M.W., SHIPLEY, G.L., VANDESOMPELE, J. and WITTWER, C.T., 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry, vol. 55, no. 4, pp. 611-622. http://dx.doi.org/10.1373/clinchem.2008.112797 PMid:19246619.
» http://dx.doi.org/10.1373/clinchem.2008.112797 - CORLESS, C.E., GUIVER, M., BORROW, R., EDWARDS-JONES, V., KACZMARSKI, E.B. and FOX, A.J., 2000. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. Journal of Clinical Microbiology, vol. 38, no. 5, pp. 47-52. http://dx.doi.org/10.1128/JCM.38.5.1747-1752.2000 PMid:10790092.
» http://dx.doi.org/10.1128/JCM.38.5.1747-1752.2000 - DAS, S., KALE, M., BEENA, P.M. and KUMAR, H., 2015. Bacterial contamination of platelet at University Hospital: “A prospective surveillance study. INSS, vol. 4, no. 4, pp. 505-508.
- DOBBELAER, I., NEERKEN, S., VAN DE WIEL, P., VAN MEERBERGEN, B., and PENTERMAN, R. Selective lysis of cells by ionic surfactants nº WO2012168003 A1. 13-12-2012.
- ECKER, D.J., SAMPATH, R., LI, H., MASSIRE, C., MATTHEWS, H.E., TOLENO, D., HALL, T.A., BLYN, L.B., ESHOO, M.W., RANKEN, R., HOFSTADLER, S.A. and TANG, Y.W., 2010. New technology for rapid molecular diagnosis of bloodstream infections. Expert Review of Molecular Diagnostics, vol. 10, no. 4, pp. 399-415. http://dx.doi.org/10.1586/erm.10.24 PMid:20465496.
» http://dx.doi.org/10.1586/erm.10.24 - ESMAILI, M.A., RAZJOU, F., KHOSROSHAHI, B.N., et al, 2017. Evaluations of detections methods of bacterial contamination in platelet components. International Journal of Medical Laboratory, vol. 4, no. 4, pp. 232-245.
- EZUKI, S., KAWABATA, K., KANNO, T. and OHTO, H., 2007. Culture-based bacterial detection systems for platelets: the effect of time prior to sampling and duration of incubation required for detection with aerobic culture. Transfusion, vol. 47, no. 11, pp. 2044-2049. http://dx.doi.org/10.1111/j.1537-2995.2007.01428.x PMid:17958533.
» http://dx.doi.org/10.1111/j.1537-2995.2007.01428.x - FISHER, R.A., 1956. Statistical methods for research workers 12th ed. London: Quarterly Journal of the Royal Meteorol Society.
- GARSON, J.A., PATEL, P., MCDONALD, C., BALL, J., ROSENBERG, G., TETTMAR, K.I., BRAILSFORD, S.R., PITT, T. and TEDDER, R.S. 2014. Evaluation of an ethidium monoazide-enhanced 16S rDNA real-time polymerase chain reaction assay for bacterial screening of platelet concentrates and comparison with automated culture. Transfusion, vol. 54, no. 3 Pt 2, pp. 870-878. http://dx.doi.org/10.1111/trf.12256 PMid:23701338.
» http://dx.doi.org/10.1111/trf.12256 - HEIN, I., SCHNEEWEISS, W., STANEK, C. and WAGNER, M., 2007. Ethidium monoazide and propidium monoazide for elimination of unspecific DNA background in quantitative universal real-time PCR. Journal of Microbiological Methods, vol. 71, no. 3, pp. 336-339. http://dx.doi.org/10.1016/j.mimet.2007.09.005 PMid:17936386.
» http://dx.doi.org/10.1016/j.mimet.2007.09.005 - HENDOLIN, P.H., MARKKANEN, A., YLIKOSKI, J. and WAHLFORS, J.J., 1997. Use of multiplex PCR for simultaneous detection of four bacterial species in middle ear effusions. Journal of Clinical Microbiology, vol. 35, no. 11, pp. 2854-2858. http://dx.doi.org/10.1128/JCM.35.11.2854-2858.1997 PMid:9350746.
» http://dx.doi.org/10.1128/JCM.35.11.2854-2858.1997 - HUMPHREY, B., MCLEOD, N., TURNER, C., SUTTON, J.M., DARK, P.M. and WARHURST, G., 2015. Removal of contaminant DNA by combined UV-EMA treatment allows low copy number detection of clinically relevant bacteria using pan-bacterial real-time PCR. PLoS One, vol. 10, no. 7, pp. e0132954. http://dx.doi.org/10.1371/journal.pone.0132954 PMid:26172943.
» http://dx.doi.org/10.1371/journal.pone.0132954 - KLAUSEGGER, A., HELL, M., BERGER, A., ZINOBER, K., BAIER, S., JONES, N., SPERL, W. and KOFLER, B., 1999. Gram type-specific broad-range PCR amplification for rapid detection of 62 pathogenic bacteria. Journal of Clinical Microbiology, vol. 37, no. 2, pp. 464-466. http://dx.doi.org/10.1128/JCM.37.2.464-466.1999 PMid:9889245.
» http://dx.doi.org/10.1128/JCM.37.2.464-466.1999 - KUMAR, S., STECHER, G. and TAMURA, K., 2016. MEGA 7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, vol. 33, no. 7, pp. 1870-1874. http://dx.doi.org/10.1093/molbev/msw054 PMid:27004904.
» http://dx.doi.org/10.1093/molbev/msw054 - LOONEN, A. J. M., BOS, M. P., VAN MEERBERGEN, B., NEERKEN, S., CATSBURG, A., DOBBELAER, I., PENTERMAN, R., MAERTENS, G., VAN DE WIEL, P., SAVELKOUL, P., and VAN DEN BRULE, A. J. C., 2013. Comparison of pathogen DNA isolation methods from large volumes of whole blood to improve molecular diagnosis of bloodstream infections. Plos One, vol. 8, no. 8, pp. e72349.
- MACAULEY, A., CHANDRASEKAR, A., GEDDIS, G., MORRIS, K.G. and MCCLELLAND, W.M., 2003. Operational feasibility of routine bacterial monitoring of platelets. Transfusion Medicine (Oxford, England), vol. 13, no. 4, pp. 189-195. http://dx.doi.org/10.1046/j.1365-3148.2003.00441.x PMid:12880389.
» http://dx.doi.org/10.1046/j.1365-3148.2003.00441.x - MANCINI, N., CARLETTI, S., GHIDOLI, N., CICHERO, P., BURIONI, R. and CLEMENTI, M., 2010. The era of molecular and other non-culture based methods in diagnosis of sepsis. Clinical Microbiology Reviews, vol. 23, no. 1, pp. 235-251. http://dx.doi.org/10.1128/CMR.00043-09 PMid:20065332.
» http://dx.doi.org/10.1128/CMR.00043-09 - MOHAMMADI, T., PIETERSZ, R.N., VANDENBROUCKE-GRAULS, C.M., SAVELKOUL, P.H. and REESINK, H.W., 2005. Detection of bacteria in platelet concentrates: comparison of broad-range real-time 16S rDNA polymerase chain reaction and automated culturing. Transfusion, vol. 45, no. 5, pp. 731-736. http://dx.doi.org/10.1111/j.1537-2995.2005.04258.x PMid:15847662.
» http://dx.doi.org/10.1111/j.1537-2995.2005.04258.x - MOHAMMADI, T., SAVELKOUL, P.H., PIETERSZ, R.N. and REESINK, H.W., 2006. Applications of real-time PCR in the screening of platelet concentrates for bacterial contamination. Expert Review of Molecular Diagnostics, vol. 6, no. 6, pp. 865-872. http://dx.doi.org/10.1586/14737159.6.6.865 PMid:17140373.
» http://dx.doi.org/10.1586/14737159.6.6.865 - PALAVECINO, E. and YOMTOVIAN, R., 2003. Risk and prevention of transfusion-related sepsis. Current Opinion in Hematology, vol. 10, no. 6, pp. 434-439. http://dx.doi.org/10.1097/00062752-200311000-00007 PMid:14564174.
» http://dx.doi.org/10.1097/00062752-200311000-00007 - PALAVECINO, E.L., YOMTOVIAN, R.A. and JACOBS, M.R., 2006. Detecting bacterial contamination in platelet products. Clinical Laboratory, vol. 56, no. 9-10, pp. 443-456. PMid:17078471.
- PAOLUCCI, M., LANDINI, M.P. and SAMBRI, V., 2010. Conventional and molecular techniques for the early diagnosis of bacteraemia. International Journal of Antimicrobial Agents, vol. 36, suppl. 2, pp. S6-S16. http://dx.doi.org/10.1016/j.ijantimicag.2010.11.010 PMid:21129933.
» http://dx.doi.org/10.1016/j.ijantimicag.2010.11.010 - PATEL, P., GARSON, J.A., TETTMAR, K.I., ANCLIFF, S., MCDONALD, C., PITT, T., COELHO, J. and TEDDER, R.S., 2012. Development of an ethidium monoazide-enhanced internally controlled universal 16S rDNA real-time polymerase chain reaction assay for detection of bacterial contamination in platelet concentrates. Transfusion, vol. 52, no. 7, pp. 1423-1432. http://dx.doi.org/10.1111/j.1537-2995.2011.03484.x PMid:22188457.
» http://dx.doi.org/10.1111/j.1537-2995.2011.03484.x - PIETERSZ, R.N., ENGELFRIET, C.P. and REESINK, H.W., 2003. Detection of bacterial contamination of platelet concentrates. Vox Sanguinis, vol. 85, no. 3, pp. 224-239. PMid:14516456.
- RAZJOU, F., NAGHADEH, H.T., FERDOWSI, S. and DABIRMOGHADAM, A., 2017. Evaluation of the sensitivity and specificity of use of glucose and pH for bacterial screening of platelet concentrates compared to the Bact/Alert. Indian Journal of Hematology & Blood Transfusion : An Official Journal of Indian Society of Hematology and Blood Transfusion, vol. 33, no. 1, pp. 116-120. http://dx.doi.org/10.1007/s12288-016-0660-4 PMid:28194067.
» http://dx.doi.org/10.1007/s12288-016-0660-4 - ROOD, I.G., PETTERSSON, A., SAVELKOUL, P.H. and DE KORTE, D., 2011. Performance and suitability of polymerase chain reaction for early detection of bacteria in platelet concentrates. Transfusion, vol. 51, no. 9, pp. 2006-2011. http://dx.doi.org/10.1111/j.1537-2995.2011.03090.x PMid:21392020.
» http://dx.doi.org/10.1111/j.1537-2995.2011.03090.x - RUECKERT, A. and MORGAN, H.W., 2007. Removal contaminating DNA from polymerase chain reaction using ethidium monoazide. Journal of Microbiological Methods, vol. 68, no. 3, pp. 596-600. http://dx.doi.org/10.1016/j.mimet.2006.11.006 PMid:17187883.
» http://dx.doi.org/10.1016/j.mimet.2006.11.006 - SCHREZENMEIER, H., WALTHER-WENKE, G., MULLER, T.H., WEINAUER, F., YOUNIS, A., HOLLAND-LETZ, T., GEIS, G., ASMUS, J., BAUERFEIND, U., BURKHART, J., DEITENBECK, R., FÖRSTEMANN, E., GEBAUER, W., HÖCHSMANN, B., KARAKASSOPOULOS, A., LIEBSCHER, U.M., SÄNGER, W., SCHMIDT, M., SCHUNTER, F., SIREIS, W. and SEIFRIED, E., 2007. Bacterial contamination of platelet concentrates: results of a prospective multicenter study comparing pooled whole blood-derived platelets and apheresis platelets. Transfusion, vol. 47, no. 4, pp. 644-652. http://dx.doi.org/10.1111/j.1537-2995.2007.01166.x PMid:17381623.
» http://dx.doi.org/10.1111/j.1537-2995.2007.01166.x - TAKAHASHI, H., YAMAZAKI, H., AKANUMA, S., KANAHARA, H., SAITO, T., CHIMURO, T., KOBAYASHI, T., OHTANI, T., YAMAMOTO, K., SUGIYAMA, S. and KOBORI, T., 2014. Preparation of Phi29 DNA Polymerase free of Amplifiable DNA using Ethidium Monoazide, an ultraviolet-free light-emitting diode lamp and trehalose. PLoS One, vol. 9, no. 2, pp. e82624. http://dx.doi.org/10.1371/journal.pone.0082624 PMid:24505243.
» http://dx.doi.org/10.1371/journal.pone.0082624 - TRUNG, N.T., HIEN, T.T.T., HUYEN, T.T.T., QUYEN, D.T., VAN SON, T., HOAN, P.Q., PHUONG, N.T., LIEN, T.T., BINH, M.T., VAN TONG, H., MEYER, C.G., VELAVAN, T.P. and SONG, H., 2016. Enrichment of bacterial DNA for the diagnosis of blood stream infections. BMC Infectious Diseases, vol. 16, no. 1, pp. 235. http://dx.doi.org/10.1186/s12879-016-1568-1 PMid:27246723.
» http://dx.doi.org/10.1186/s12879-016-1568-1 - VAN MEERBERGEN, B.E.G.J., PICIU, O.M., GIL, R., SCHMIDT, K.A., NEERKEN, S., PONJEE, M.W.G., UNAY, Z.S., PENTERMAN, R. and VAN DE WIEL, P.A. Selective lysis of cell nº WO2011070507 A1. 16-06-2011.
- WAGNER, S.J., 2004. Transfusion-transmitted bacterial infection: Risks, sources and interventions. Vox Sanguinis, vol. 86, no. 3, pp. 157-163. http://dx.doi.org/10.1111/j.0042-9007.2004.00410.x PMid:15078249.
» http://dx.doi.org/10.1111/j.0042-9007.2004.00410.x - WALTHER-WENKE, G., 2008. Incidence of bacterial transmission and transfusion reactions by blood components. Clinical Chemistry and Laboratory Medicine, vol. 46, no. 7, pp. 919-925. http://dx.doi.org/10.1515/CCLM.2008.151 PMid:18605950.
» http://dx.doi.org/10.1515/CCLM.2008.151 - WILSON, K.H., BLITCHINGTON, R.B. and GREENE, R.C., 1990. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. Journal of Clinical Microbiology, vol. 28, no. 9, pp. 1942-1946. http://dx.doi.org/10.1128/JCM.28.9.1942-1946.1990 PMid:2095137.
» http://dx.doi.org/10.1128/JCM.28.9.1942-1946.1990 - WORLD HEALTH ORGANIZATION – WHO, 2009. CDC protocol of real time RTPCR for swine influenza A(H1N1) Geneva: WHO, pp. 1-9.
- YANG, S., LIN, S., KELEN, G.D., QUINN, T.C., DICK, J.D., GAYDOS, C.A. and ROTHMAN, R.E., 2002. Quantitative multiprobe PCR assay for simultaneous detection and identification to species level of bacterial pathogens. Journal of Clinical Microbiology, vol. 40, no. 9, pp. 3449-3454. http://dx.doi.org/10.1128/JCM.40.9.3449-3454.2002 PMid:12202592.
» http://dx.doi.org/10.1128/JCM.40.9.3449-3454.2002
Publication Dates
-
Publication in this collection
14 Aug 2020 -
Date of issue
Jul-Sep 2021
History
-
Received
11 Oct 2019 -
Accepted
08 Mar 2020