Abstract
Enteropathogenic Escherichia coli (EPEC) comprise one of the six categories of diarrhoeagenic E. coli (DEC). EPEC is subgrouped into typical (tEPEC) and atypical (aEPEC). The identification of DEC cannot be based only on cultural and biochemical criteria, since they are indistinguishable from the non-pathogenic E. coli commonly found in human feces. Several PCR methods, with both single and multiple target genes, have been reported for detecting the different DEC pathotypes. In the present study five hundred E. coli isolates from children with diarrhea were subjected into multiplex PCR. Furthermore the strains were typed serologically with O antisera and their fliC gene was characterized by PCR-RFLP. The results obtained revealed that overall 41 (8.2%) isolates could be detected as EPEC by this multiplex PCR assay. Of these isolates; 27 (66%) were typical (escv+, bfp+) and 14 (34%) atypical EPEC (escv+, bfp-). None of these 41 isolates contained the Stx1 and Stx2 genes. Among 37 (90%) typeable strains, nine different serogroups were present. The most common serogroups were O111, followed by O86, O55 and O119 and 10 different H types were found among these isolates. The multiplex PCR assay was found to be rapid and reliable in comparison to serological test; especially when screening the large number of isolates.
diarrhea; enteropathogenic Escherichia coli; polymerase chain reaction
ORIGINAL ARTICLE
Comparison of multiplex PCR with serogrouping and PCR-RFLP of fliC gene for the detection of enteropathogenic Escherichia coli (EPEC)
Saeid BouzariI; Mohammad M AslaniII; Mana OloomiI; Anis JafariI; Amir DashtiIII
IPhD, Researcher, Molecular Biology Unit, Pasteur Institute of Iran, Tehran, Iran
IIPhD, Researcher, Bacteriology Department, Pasteur Institute of Iran, Tehran, Iran
IIIMSc, Researcher, Molecular Biology Unit, Pasteur Institute of Iran, Tehran, Iran
Correspondence to Correspondence to: Saeid Bouzari Pasteur Institute of Iran, Molecular Biology Unit Tehran, Iran 1316943551 Phone: +9821 6695331120 (ext: 2223) Fax: +9821 66492619 saeidbouzari@yahoo.com
ABSTRACT
Enteropathogenic Escherichia coli (EPEC) comprise one of the six categories of diarrhoeagenic E. coli (DEC). EPEC is subgrouped into typical (tEPEC) and atypical (aEPEC). The identification of DEC cannot be based only on cultural and biochemical criteria, since they are indistinguishable from the non-pathogenic E. coli commonly found in human feces. Several PCR methods, with both single and multiple target genes, have been reported for detecting the different DEC pathotypes. In the present study five hundred E. coli isolates from children with diarrhea were subjected into multiplex PCR. Furthermore the strains were typed serologically with O antisera and their fliC gene was characterized by PCR-RFLP. The results obtained revealed that overall 41 (8.2%) isolates could be detected as EPEC by this multiplex PCR assay. Of these isolates; 27 (66%) were typical (escv+, bfp+) and 14 (34%) atypical EPEC (escv+, bfp-). None of these 41 isolates contained the Stx1 and Stx2 genes. Among 37 (90%) typeable strains, nine different serogroups were present. The most common serogroups were O111, followed by O86, O55 and O119 and 10 different H types were found among these isolates. The multiplex PCR assay was found to be rapid and reliable in comparison to serological test; especially when screening the large number of isolates.
Keywords: diarrhea; enteropathogenic Escherichia coli; polymerase chain reaction.
INTRODUCTION
Diarrhea is an important public health problem with high infant mortality especially in developing countries. Systematic surveys have shown that diarrhoeagenic Escherichia coli (DEC) are a common cause of diarrhea in both developing and developed countries. To date six categories of DEC have been defined on the basis of specific virulence properties.1,2 Enterotoxigenic E. coli (ETEC) cause diarrhea via secretion of heat stable (ST) and/or heat labile (LT) enterotoxin. Enteroinvasive E. coli (EIEC) strains are closely related to Shigella spp. and the responsible genes are carried on the pINV plasmid. Shiga-toxin producing E. coli (STEC) or enterohaemorrhagic non-bloody diarrhea as well as a hemorrhagic colitis may trigger haemolytic uraemic syndrome (HUS).
Infant diarrhea in developing countries is linked to EPEC strains,1 but they are also found with increasing frequency from diarrheal cases in industrialized countries.2 The ability to induce attaching-effacing (A/E) lesions in the intestine is main characteristic of EPEC.3-5 The genes required for the production of these lesions are located on a pathogenicity island known as the locus for enterocyte effacement (LEE). The eae and escV genes of EPEC encode factors responsible for the attaching and effacing lesions that intimate bacterial adherence to the intestinal epithelial cells. These lesions are comprised of bacteria intimately attached to the intestinal mucosa at sites of cytoskeletal rearrangements leading to characteristic morphological changes, known as cupping and pedestal formation, accompanied by the absence of microvilli.6
EPEC is divided into two groups: typical and atypical. The basic difference between the typical EPEC (tEPEC) and atypical EPEC (aEPEC) is a ca. 90-kb EPEC adherence factor plasmid (pEAF) that encodes type IV-like bundle-forming pili (BFP), whereas atypical EPEC lacks this adherence promoting factor.7 The tEPEC produce localized adherence to epithelial cells in cultures forming microcolonies on the cell surface.8,9 Adult volunteers have demonstrated that these genes are essential virulence determinants of EPEC.10-12 The involvement of some aEPEC strains with diarrheal outbreak support the concept that they are also pathogenic.2
Since DEC cannot be diagnosed adequately by culture and biochemical criteria alone, identification of these strains is difficult. Traditional methods to detect diarrheagenic E. coli have been very laborious and not suitable for daily clinical use, except for EHEC. Therefore, systematic epidemiological data of diarrheagenic E. coli prevalence in different patient groups have not been available either.13 To overcome this obstacle, molecular methods detecting all major diarrheagenic E. coli have recently been described. Many PCR techniques detecting various genes coding for virulence traits of the different categories of DEC have been reported.14-19 In this study we screened large number of clinical isolates of E. coli by multiplex PCR.20 The results of PCR assay was compared with traditional typing, i.e., serology for O typing and PCR-RFLP technique for H typing of the isolates. Therefore the main features of these techniques are presented and discussed in this present communication.
MATERIAL AND METHODS
Bacterial strains
In this study 500 E. coli isolates from diarrheal patients were subjected to multiplex PCR and PCR- RFLP and serological typing. E. coli strains E2348/69 were used as positive control.
DNA extraction
The genomic DNA of the isolates was extracted with alkaline lysis method21 and was used for the PCR purpose. The method22 was followed for total genomic DNA extraction that is used for PCR-RFLP.
Multiplex PCR
Molecular typing was performed using multiplex PCR that can identify STEC and EPEC pathogroups based on sequences complementary to escV, bfpB, Stx1, and Stx2.
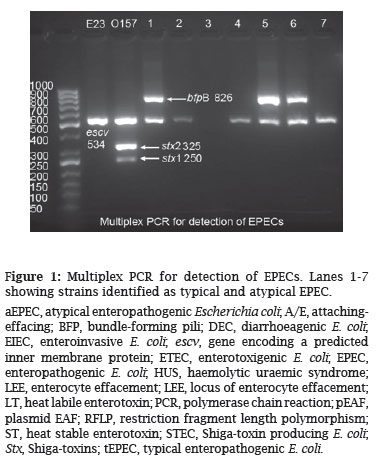
This multiplex PCR differentiates STEC and EPEC and also divides the later into two groups, i.e. typical and atypical EPEC. Strains with escv+, bfp+ traits are considered typical and isolates with escv+, bfp- are atypical EPEC. The PCR was performed in a 25 µL reaction mixture consisting of 1 U of Taq DNA polymerase with the corresponding Taq polymerase buffer, a 0.3 mM concentration of each deoxynucleoside triphosphate, and a 0.4 mM concentration of each PCR primer. Thermocycling conditions were as follows: 95ºC for 5 min, followed by 30 cycles of 95ºC for 1 min, 58ºC for 1 min, and 72ºC for 1 min, with a final extension at 72ºC for 5 min. The sequence of primers and expected PCR product with the appropriate size are shown in Table 1. The PCR products hence obtained were run on 1% gel and the gels were visualized by gel Doc system (Figure 1).
O typing
The O antigens of the isolates were typed with commercially available "O" antisera with slide agglutination.
fliC gene amplification
The PCR-RFLP technique23 was employed for determination of H type of the isolates. For this purpose the fliC gene of the isolates was amplified using specific primers; the sequence of primers and PCR condition is as follows:
Hot start: 94ºC 5 min
Denaturation: 94ºC 1 min
Annealing: 55ºC 1 min
Extension: 72ºC 1 min
Final extension: 72ºC 5 min
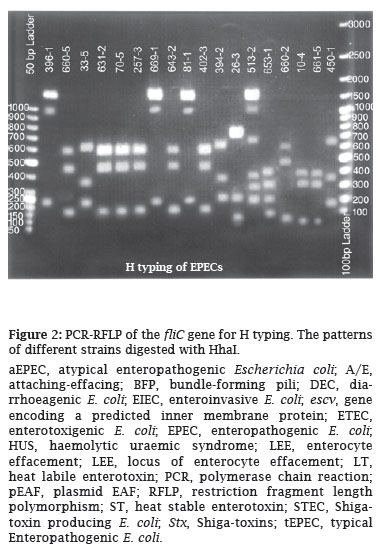
The PCR products obtained were eluted from the gel and digested with Hha1 and the digested patterns of the isolates were compared and thereafter the H types of the isolates were determined (Figure 2).
RESULTS
In the present study a multiplex PCR was employed for the detection of typical and atypical EPEC among 500 E. coli isolates. The results obtained revealed that overall 41(8.2%) isolates could be detected as EPEC by this multiplex PCR assay. Of these isolates; 27 (66%) were typical (escv+, bfp+) and 14 (34%) atypical EPEC (escv+, bfp-) (Figure 1). The isolates were then subjected to serogrouping and nine different serogroups were detected. On the other hand, strains typeable with O antisera were 37 (90.2%) and 4 (9.8%) were found to be non-typeable with O antisera. Among typeable strains, nine different serogroups were present as follows: O111, O86, O55, O119, O127, O126, O114, O142, and O26. The most common serogroup was O111 followed by O86, O55, each by 7, 6 and 5 strains respectively. All these serogroups were distributed among typical and atypical EPEC, and only among strains with serogroups O114, O126 and O127 all the isolates were found to be typical EPEC by PCR analysis. For the rest of the serogroups typical and atypical isolates were distributed randomly. Of the 37 typeable strains, 25 (67.6%) were typical and 12 (32.4%) atypical and for 4 non-typeable isolates 2 were typical and the rest 2 were atypical. H typing with PCR-RFLP revealed 10 different H types, being the most common H types H41, H2 and H7 (each with 8, 7 and 5 isolates, respectively - Figure 2). Overall in 37 typeable strains, 11 (29.7%) strains could not have the H determined, and for 4 non-typeable strains the H types, was determined. On the basis of O:H typing, 26 strains (5.2%) could be fully characterized (Table 2).
DISCUSSION
Infant diarrhea in developing countries is linked to EPEC strains,1 but they are also found with increasing frequency from diarrheal cases in industrialized countries. In 1995, EPEC was divided into two groups, typical EPEC (tEPEC) and atypical EPEC (aEPEC). The basic difference between the two groups is the presence of the EPEC adherence factor plasmid (pEAF) in tEPEC and its absence in aEPEC.2
The World Health Organization recognized that EPEC comprises strains of 12 O serogroups: O26, O55, O86, O111, O114, O119, O125, O126, O127, O128, O142 and O158,24 also known as the classical EPEC serogroups. Identification of EPEC and STEC strains is currently usually based on serotyping with specific antisera in a time-consuming process demanding some technical expertise.20 In the present investigation, the traditional method of serotyping targeting exclusively the 12 serogroups considered to represent the EPEC pathotype was performed. Only 37 (7.4%) strains were found to be typeable with O antisera. The PCR-RFLP was performed for H typing of the isolates in conjunction with O typing and with combination of O:H typing 26 strains (5.2%) could be fully characterized.
Since DEC cannot be diagnosed adequately by culture and biochemical criteria alone, identification of these strains is difficult. Traditional methods to detect diarrheagenic E. coli have been very laborious and not suitable for daily clinical use. To facilitate diagnostic and therapeutic measures, many PCR techniques detecting various genes coding for virulence traits of the different categories of DEC have been reported.14-20 Several PCR assays have been developed for rapid identification and differentiation of diarrheagenic E. coli is belonging to EPEC and STEC pathotypes. The study of Abe et al.25 from Brazil indicated that aEPEC comprise a very heterogeneous group. Typing of O and H antigens showed a large variety of serotypes (52 O:H combinations), including the O non-typeable strains. Most of the strains belonged to serotypes not included in classical EPEC serogroups, and several strains were non-motile. EHEC strains cause acute inflammation in the intestine and bloody diarrhea, including abdominal pain with, rare but severe, secondary complications such as haemolytic uremic syndrome (HUS). The main virulence factors of EHEC are Shiga-toxins (Stx) that induce cytotoxic effects on the microvascular endothelial cells.10 The PCR technique of Muller et al.20 used in the present study is a novel single MPCR that allows for the specific differential detection of LEE-harboring EPEC, ATEC, and STEC isolates and also identifies LEE-negative STEC strains in a straightforward and robust reaction. The eight primers developed by Muller et al.20 proved to be specific for the corresponding four genes and, its evaluation with large number of clinical E. coli isolates demonstrated that the MPCR is highly specific and reliable. It was conclude that the MPCR is a specific method for the identification and differentiation of EPEC, ATEC, STEC, and LEE-negative STEC strains and might be as routine diagnostic tool. In our study none of the isolates were found to be Stx producer, this could be due to absence of Stx producer isolates from our collection or they are Stx-producing E. coli (STEC) that have lost the toxin-encoding prophage. Although the epidemiological association of aEPEC with diarrhea is still controversial, its high prevalence worldwide and the involvement of some strains with diarrheal outbreaks support the concept that some aEPEC strains are diarrheagenic. However the findings of Tennant et al.26 indicated that clinical isolates of aEP-EC obtained from patients in Australia or New Zealand are not derived from tEPEC or STEC and they probably carry other virulence determinants that could compensate for their lack of BFP. Although in a study conducted by Alikhani et al.27 atypical EPEC isolation was reported from diarrheal cases. Two hundred and twelve fecal samples collected from 62 chickens, 50 ducks and 100 pigeons were investigated for the presence of Stx1, Stx2, eae and ehxA virulence genes by multiplex PCR. All EPEC isolates were atypical as they lacked bfpA and atypical EPEC were from poultry.28 However, the 14 isolates detected as (escv+, bfpA-) atypical EPEC by this PCR assay in our study, could either represent LEE-positive (esciv+) atypical isolates or tEPEC that have lost the genes for bfp (bfpA-). Comparing the isolates detected by PCR (8.2%) with O typed strains (7.4%) and O:H typed strains (5.2%); it is obvious that PCR is more appropriate technique especially when large numbers of isolates are considered for detection. Overall the results obtained in the present study revealed that the multiplex PCR assay is useful tool for the rapid screening of the large number of isolates. It is very appropriate technique in an epidemic situation where the rapid diagnosis is required. However use of serotyping is necessary in surveillance studies when more information regarding the isolates is required.
ACKNOWLEDGEMENTS
This project was financially supported by Pasteur Institute of Iran (grant no: 268).
Submitted on: 10/10/2010
Approved on: 11/17/2010
We declare no conflict of interest.
- 1. Robins-Browne RM. Traditional enteropathogenic Escherichia coli of infantile diarrhea. Rev Infect Dis 1987;9(1):28-53.
- 2. Trabulsi LR, Keller R, Tardelli Gomes TA. Typical and atypical enteropathogenic Escherichia coli Emerg Infect Dis 2002;8(5):508-13.
- 3. Moon HW, Whipp SC, Argenzio RA, Levine MM et al. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect Immun 1983;41(3):1340-51.
- 4. Rothbaum RJ, Partin JC, Saalfield K et al. An ultrastructural study of enteropathogenic Escherichia coli infection in human infants. Ultrastruct Pathol 1983;4(4):291-304.
- 5. Tzipori S, Robins-Browne RM, Gonis G et al. Enteropathogenic Escherichia coli enteritis: evaluation of the gnotobiotic piglet as a model of human infection. Gut 1985;26(6):570-8.
- 6. Celli J, Deng W, Finlay BB. Enteropathogenic Escherichia coli (EPEC) attachment to epithelial cells: exploiting the host cell cytoskeleton from the outside. Cell Microbiol 2000;2(1):1-9.
- 7. Tobe T, Hayashi T, Han CG et al. Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coli adherence factor plasmid. Infect Immun 1999;67(10):5455-62.
- 8. Cleary J, Lai LC, Shaw RK et al. Enteropathogenic Escherichia coli (EPEC) adhesion to intestinal epithelial cells: role of bundle-forming pili (BFP), EspA filaments and intimin. Microbiology 2004;150(Pt 3):527-38.
- 9. Tobe T, Sasakawa C. Role of bundle-forming pilus of enteropathogenic Escherichia coli in host cell adherence and in microcolony development. Cell Microbiol 2001;3(9):579-85.
- 10. Bieber D, Ramer SW, Wu CY et al. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli Science 1998;280(5372):2114-8.
- 11. Donnenberg MS, Tacket CO, James SP et al. Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J Clin Invest 1993;92(3):1412-7.
- 12. Levine MM, Nataro JP, Karch H et al. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J Infect Dis1985;152(3):550-9.
- 13. Antikainen J, Tarkka E, Haukka K et al. New 16-plex PCR method for rapid detection of diarrheagenic Escherichia coli directly from stool samples. Eur J Clin Microbiol Infect Dis 2009;28(8):899-908.
- 14. Aranda KR, Fagundes-Neto U, Scaletsky IC. Evaluation of multiplex PCRs for diagnosis of infection with diarrheagenic Escherichia coli and Shigella spp. J Clin Microbiol 2004;42(12):5849-53.
- 15. Brandal LT, Lindstedt BA, Aas L et al. Octaplex PCR and fluorescence-based capillary electrophoresis for identification of human diarrheagenic Escherichia coli and Shigella spp. J Microbiol Methods 2007;68(2):331-41.
- 16. Kimata K, Shima T, Shimizu M et al. Rapid categorization of pathogenic Escherichia coli by multiplex PCR. Microbiol Immunol 2005;49(6):485-92.
- 17. Pass MA, Odedra R, Batt RM. Multiplex PCRs for identification of Escherichia coli virulence genes. J Clin Microbiol 2000;38(5):2001-4.
- 18. Vidal M, Kruger E, Durán C et al. Single multiplex PCR assay to identify simultaneously the six categories of diarrheagenic Escherichia coli associated with enteric infections. J Clin Microbiol 2005;43(10):5362-5.
- 19. Hardegen C, Messler S, Henrich B et al. A set of novel multiplex Taqman real-time PCRs for the detection of diarrhoeagenic Escherichia coli and its use in determining the prevalence of EPEC and EAEC in a university hospital. Ann Clin Microbiol Antimicrob 2010;9:5-7.
- 20. Müller D, Hagedorn P, Brast S et al. Rapid identification and differentiation of clinical isolates of enteropathogenic Escherichia coli (EPEC), atypical EPEC, and Shiga toxin-producing Escherichia coli by a one-step multiplex PCR method. J Clin Microbiol 2006;44(7):2626-9.
- 21. Sambrook J, Russell DW. Molecular cloning, Cold Spring Harbor Laboratory Press, 2001; Cold Spring Harbor, New York, p. 1.32-1.34.
- 22. Owen RJ, Borman P. A rapid biochemical method for purifying high molecular weight bacterial chromosomal DNA for restriction enzyme analysis. Nucleic Acids Res 1987;15(8):3631.
- 23. Machado J, Grimont F, Grimont PA. Identification of Escherichia coli flagellar types by restriction of the amplified fliC gene. Res Microbiol 2000;151(7):535-46.
- 24. World Health Organization Programme for Control of Diarrhoeal Diseases. Manual for Laboratory Investigations of Acute Enteric Infections. 1987; World Health Organization, Geneva.
- 25. Abe CM, Trabulsi LR, Blanco J et al. Virulence features of atypical enteropathogenic Escherichia coli identified by the eae(+) EAF-negative stx(-) genetic profile. Diagn Microbiol Infect Dis 2009;64(4):357-65.
- 26. Tennant SM, Tauschek M, Azzopardi K et al. Characterization of atypical enteropathogenic E. coli strains of clinical origin. BMC Microbiol 2009;9:117.
- 27. Alikhani MY, Mirsalehian A, Aslani MM. Detection of typical and atypical enteropathogenic Escherichia coli (EPEC) in Iranian children with and without diarrhoea. J Med Microbiol 2006;55(Pt9):1159-63.
- 28. Farooq S, Hussain I, Mir MA et al. Isolation of atypical enteropathogenic Escherichia coli and Shiga toxin 1 and 2f-producing Escherichia coli from avian species in India. Lett Appl Microbiol 2009;48(6):692-7.
Correspondence to:
Publication Dates
-
Publication in this collection
15 Aug 2011 -
Date of issue
Aug 2011
History
-
Received
10 Oct 2010 -
Accepted
17 Nov 2010