Abstract:
The objective of this work was to evaluate the evolutionary dynamics of the wild-weedy-domestic gene pool of Cucurbita argyrosperma squash by estimating the levels of genetic diversity and gene flow in the putative area of its domestication. Nine populations were collected, and DNA was extracted from young leaves harvested separately from approximately 20 individuals in each population. The DNA fragments were amplified with eight pairs of SSR primers and separated by electrophoresis in 5% denaturing polyacrylamide gels. Genetic diversity and the amount of gene flow were estimated in the populations, and Bayesian grouping was used to determine the levels of gene infiltration and probability of ancestry. The ethnobotanical exploration indicated that the evolutionary dynamics in the area occurred under five different ecological scenarios. Eighty-seven alleles with 75% to 100% polymorphic loci were identified. The greater genetic diversity in the weedy-domestic populations may have been the product of recombination due to the high gene flow between these populations promoted by pollinators and human selection. There is high gene flow between the wild and cultivated populations of C. argyrosperma in its domestication centre, highlighting the importance of conserving and maintaining these genetic resources.
Index terms:
Cucurbitaceae; genetic diversity; microsatellite markers; plant genetic resources; squash
Resumo:
O objetivo deste trabalho foi avaliar a dinâmica evolutiva do pool genético silvestre-arvense-doméstico da abóbora (Cucurbita argyrosperma) ao estimar os níveis de diversidade genética e o fluxo gênico em sua provável área domesticação. Foram coletadas nove populações, tendo-se extraído o DNA de folhas jovens de cerca de 20 plantas de cada população. Os fragmentos de DNA foram amplificados por meio de oito pares de iniciadores de sequência simples repetida (SSR) e separados por eletroforese em 5% de gel de desnaturação de poliacrilamida. Estimaram-se a diversidade genética e o fluxo gênico para as populações, e utilizou-se o agrupamento bayesiano para determinar os níveis de infiltração gênica e probabilidade de ancestralidade. A investigação etnobotânica mostrou que a dinâmica evolutiva na área ocorreu em cinco cenários ecológicos diferentes. Foram identificados 87 alelos com 75 e 100% de loci polimórficos. A maior diversidade genética nas populações arvense-domésticas pode ter sido resultante da recombinação devido ao alto fluxo gênico entre estas populações, ocasionado pelos polinizadores e pela seleção humana. Há grande fluxo gênico entre as populações silvestres e cultivadas de C. argyrosperma no centro de domesticação, o que indica a importância da conservação e da manutenção destes recursos genéticos.
Termos para indexação:
Cucurbitaceae; diversidade genética; marcadores microssatélites; recursos fitogenéticos; abóbora
Introduction
Mesoamerica is considered the center of origin and diversity of the genus Cucurbita. Fifteen of the 20 taxa that flourish were possibly domesticated within this territory, since the wild ancestors of the species C. argyrosperma subsp. argyrosperma and C. pepo subsp. fraterna are distributed in this area (Lira et al., 2016LIRA, R.; EGUIARTE L.; MONTES, S.; ZIZUMBO-VILLARREAL, D.; COLUNGA-GARCÍAMARIN, P.; QUESADA, M. Homo sapiens-Cucurbita interaction in Mesoamerica: domestication, dissemination, and diversification. In: LIRA, R.; CASAS, A.; BLANCAS, J. (Ed.). Ethnobotany of Mexico: interactions of people and plants in Mesoamerica. New York: Springer, 2016. p.389-400. DOI: 10.1007/978-1-4614-6669-7_15.
https://doi.org/10.1007/978-1-4614-6669-...
). It was in the Mesoamerican region that species such as maize (Zea mays L.), beans (Phaseolus spp.), and squash (Cucurbita spp.) were domesticated and integrated into a multi-crop system known in the region as milpa (Zizumbo-Villarreal & Colunga-GarcíaMarín, 2010ZIZUMBO-VILLARREAL, D.; COLUNGA-GARCÍAMARÍN, P. Origin of agriculture and plant domestication in West Mesoamerica. Genetic Resources and Crop Evolution, v.57, p.813-825, 2010. DOI: 10.1007/s10722-009-9521-4.
https://doi.org/10.1007/s10722-009-9521-...
).
It has been suggested that the biogeographic region of Balsas-Jalisco, Mexico, could be the center of domestication of C. argyrosperma subsp. argyrosperma, as this is an area where the species continues to be cultivated and where both wild and domestic populations continue to provide seeds for consumption under pre-Columbian food processing techniques. Archaeological studies indicate that the domestication of C. argyrosperma could have occurred 8,900 years ago, as this species is a floristic component of the low deciduous forest that settlers made into food together with maize kernels (Sanjur et al., 2002SANJUR, O.I.; PIPERNO, D.R.; ANDRES, T.C.; WESSEL-BEAVER, L. Phylogenetic relationships among domesticated and wild species of Cucurbita (Cucurbitaceae) inferred from a mitochondrial gene: implications for crop plant evolution and areas of origin. Proceedings of the National Academy Sciences of the United States of America, v.99, p.535-540, 2002. DOI: 10.1073/pnas.012577299.
https://doi.org/10.1073/pnas.012577299....
; Zizumbo-Villarreal et al., 2012ZIZUMBO-VILLARREAL, D.; FLORES-SILVA, A.; COLUNGA-GARCÍA MARÍN, P. The Archaic diet in Mesoamerica: incentive for milpa development and species domestication. Economic Botany, v.66, p.328-343, 2012. DOI: 10.1007/s12231-012-9212-5.
https://doi.org/10.1007/s12231-012-9212-...
). In milpa, the traditional agricultural system of western Mesoamerica, it is common to find wild and domestic populations of maize (Zea mays subsp. parviglumis), beans (Phaseolus spp.), and squash (Cucurbita argyrosperma subsp. sororia L.H. Bailey) growing together in crop fields, on fallow land or in disturbed ruderal areas (Merrick, 1990MERRICK, L.C. Systematics and evolution of a domesticated squash, Cucurbita argyrosperma, and its wild and weedy relatives. In: BATES, D.M.; ROBINSON, R.W.; JEFFREY, C. (Ed.). Biology and utilization of the Cucurbitaceae. Ithaca: Cornell University Press, 1990. p.77-95.; Zizumbo-Villarreal et al., 2014ZIZUMBO-VILLARREAL, D.; FLORES-SILVA, A.; COLUNGA-GARCÍAMARÍN, P. The food system during the formative period in west Mesoamerica. Economic Botany, v.68, p.67-84, 2014. DOI: 10.1007/s12231-014-9262-y.
https://doi.org/10.1007/s12231-014-9262-...
).
Cross-fertilization can occur between domesticated, cultivated, and wild populations (Wilson et al., 1994WILSON, H.D.; LIRA, R.; RODRÍGUEZ, I. Crop/weed gene flow: Cucurbita argyrosperma Huber and C. fraterna L.H. Bailey (Cucurbitaceae). Economic Botany, v.48, p.293-300, 1994. DOI: 10.1007/BF02862330.
https://doi.org/10.1007/BF02862330....
). This shows that, it is possible to generate hybrid plants resulting from natural crosses between wild and domestic individuals and their backcrosses, which eventually forms genetically distinct populations of wild-weedy-domesticated plants. Under such conditions, there is active gene flow from the wild to the domesticated populations, as well as in the opposite direction. The resulting populations can, therefore, grow in different natural or agricultural scenarios, where different selection pressures shape the diversity and structure of the populations. These selective pressures encompass the environmental conditions under which both natural plant communities and intensive crops are grown (Payró de la Cruz et al., 2005PAYRÓ DE LA CRUZ, E.; GEPTS, P.; COLUNGA GARCIAMARÍN, P.; ZIZUMBO VILLARREAL, D. Spatial distribution of genetic diversity in wild populations of Phaseolus vulgaris L. from Guanajuato and Michoacán, México. Genetic Resources and Crop Evolution, v.52, p.589-599, 2005. DOI: 10.1007/s10722-004-6125-x.
https://doi.org/10.1007/s10722-004-6125-...
).
Studies on C. argyrosperma performed in western Mesoamerica indicate that this gene flow is promoted by floral biology, sympatry, and the presence of both native and introduced pollinators. These works also indicate that the hybrids that form in both directions produce highly fertile progeny (Lira Saade, 1995LIRA SAADE, R. Estudios taxonómicos y ecogeográficos de las cucurbitaceae latinoamericanas de importancia económica: Cucurbita, Sechium, Sicana y Cyclanthera. Rome: International Plant Genetic Resources Institute, 1995. 281p. (Systematic and Ecogeographic Studies on Crop Gene Pools, v. 9).; Montes-Hernandez & Eguiarte, 2002MONTES-HERNANDEZ, S.; EGUIARTE, L.E. Genetic structure and indirect estimates of gene flow in three taxa of Cucurbita (Cucurbitaceae) in western Mexico. American Journal of Botany, v.89, p.1156-1163, 2002. DOI: 10.3732/ajb.89.7.1156.
https://doi.org/10.3732/ajb.89.7.1156....
, Lira et al., 2016LIRA, R.; EGUIARTE L.; MONTES, S.; ZIZUMBO-VILLARREAL, D.; COLUNGA-GARCÍAMARIN, P.; QUESADA, M. Homo sapiens-Cucurbita interaction in Mesoamerica: domestication, dissemination, and diversification. In: LIRA, R.; CASAS, A.; BLANCAS, J. (Ed.). Ethnobotany of Mexico: interactions of people and plants in Mesoamerica. New York: Springer, 2016. p.389-400. DOI: 10.1007/978-1-4614-6669-7_15.
https://doi.org/10.1007/978-1-4614-6669-...
).
In this context, several questions are important in terms of the current evolution of the squash gene pool in the domestication center of the species and also of the possible in situ and ex situ conservation strategies of wild and domestic phytogenetic collections. Gene flow among wild and domesticated populations has multiple potential consequences, both positive, such as increased diversity and adaptation (Hufford et al., 2013HUFFORD, M.B.; LUBINKSY, P.; PYHÄJÄRVI, T.; DEVENGENZO, M.T.; ELLSTRAND, N.C.; ROSS-IBARRA, J. The genomic signature of crop-wild introgression in maize. Plos Genetics, v.9, e1003477, 2013. DOI: 10.1371/annotation/2eef7b5b-29b2-412f-8472-8fd7f9bd65ab.
https://doi.org/10.1371/annotation/2eef7...
), and negative, such as the unwanted escape of genes that leads to the emergence of aggressive weeds and propagation of transgenes to wild populations (Piñeyro-Nelson et al., 2009PIÑEYRO-NELSON, A.; VAN HEERWAARDEN, J.; PERALES, H.R.; SERRATOS- HERNÁNDEZ, J.A.; RANGEL, A.; HUFFORD, M.B.; GEPTS, P.; GARAY-ARROYO, A.; RIVERA-BUSTAMANTE, R.; ÁLVAREZ-BUYLLA, E.R. Transgenes in Mexican maize: molecular evidence and methodological considerations for GMO detection in landrace populations. Molecular Ecology, v.18, p.750-761, 2009. DOI: 10.1111/j.1365-294X.2008.03993.x.
https://doi.org/10.1111/j.1365-294X.2008...
).
The objective of this work was to evaluate the evolutionary dynamics of the wild-weedy-domestic gene pool of C. argyrosperma by estimating the levels of genetic diversity and gene flow in the putative area of its domestication.
Materials and Methods
The study was performed in the putative area of domestication of C. argyrosperma subsp. sororia, or the Balsas-Jalisco biogeographic region of western Mesoamerica, in the states of Jalisco and Colima between the banks of the Armería River and the Colima volcano (Table 1). The geographical coordinates of the study were 19.27-19.31N, 103.71-103.75W (Zizumbo-Villareal et al., 2009ZIZUMBO-VILLARREAL, D.; GONZÁLEZ-ZOZAYA, F.; OLAY-BARRIENTOS, A.; PLATAS-RUÍZ, R.; CUEVAS-SAGARDÍ, M.; ALMENDROS-LÓPEZ, L.; COLUNGA-GARCÍAMARÍN, P. Archaeological evidence of the cultural importance of Agave spp. in pre-hispanic Colima, Mexico. Economic Botany, v.63, p.288-302, 2009.). In 2014, nine populations of squash were collected in different agroecological scenarios at an altitudinal range of 350 to 1,150 m. Population 1 (P1) was collected from spontaneous growth beside the Armería River bed under natural conditions, which included natural disturbance caused by the rising of the river. Populations 2 and 3 (P2 and P3) were collected from spontaneous growth beside roads under conditions disturbed by human activity. Populations 4, 5, and 9 (P4, P5, and P9) were collected from spontaneous growth at sites disturbed by abandoned or long-fallow crops. Populations 6 and 8 (P6 and P8) were collected from spontaneous growth as weeds among short-fallow crops. Population 7 (P7) was domesticated and grown under cultivation associated with maize and beans in Zapotitlán, Jalisco, Mexico.
Sites in the states of Jalisco and Colima, Mexico, where the nine populations of squash (Cucurbita argyrosperma) studied with SSR markers were collected in 2014.
From each population, one fruit was collected from approximately 20 plants, and, from each fruit, a plant was grown for DNA extraction. P2, P3, P4, P5, and P6 had fewer than 20 individuals, so the total number was used (15, 19, 16, 13, and 18 individuals, respectively). Twenty pairs of Simple Sequence repeat (SSR) primers for putative Cucurbita loci were developed, and a scan was performed to select the primers with the highest sensitivity for polymorphisms. The work was performed in the laboratory of molecular markers at Centro de Investigación Científica de Yucatán, located Mérida, Mexico.
DNA was extracted from young leaves harvested separately from the individuals in each population. The extraction was performed using silica according to the method described by Echevarría-Machado et al. (2005)ECHEVARRÍA-MACHADO, I.; SÁNCHEZ-CACH, L.A.; HERNÁNDEZ-ZEPEDA, C.; RIVERA-MADRID, R.; MORENO-VALENZUELA, O.A. A simple and efficient method for isolation of DNA in high mucilaginous plant tissues. Molecular Biotechnology, v.31, p.129-135, 2005. DOI: 10.1385/MB:31:2:129.
https://doi.org/10.1385/MB:31:2:129....
. The quality and quantity of the DNA were assessed on 1% agarose gel containing 0.2 mg mL-1 ethidium bromide. From each sample, 2 mL DNA were treated with 3 μL of type IV dye (0.25% blue bromophenol and 40% sucrose). The samples were assessed using the NanoDrop 2000 UV-VIS spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA) at wavelengths of 260 and 280 nm, and the extracted DNA was diluted with sterile water to a concentration of 5 ng μL-1.
Microsatellite amplification reactions (SSR-PCR) were performed in 0.2-mL tubes containing a final reaction volume of 25 μL. The reaction mixture for each individual consisted of 5 μL of 10x PCR buffer (200 mmol L-1 Tris-HCl [pH 8.4], 500 mmol L-1 KCl), 1 μL magnesium chloride (50 mmol L-1 MgCl2), 0.5 μL of the dNTP mixture (100 mmol L-1 dATP, dTTP, dGTP, and dCTP), 1 μL of Primer F and Primer R, 0.3 μL Taq polymerase, 2 μL genomic DNA (5 ng), and 14.2 μL sterile water to reach the final volume. Amplifications were performed on the GeneAmp PCR System 9700 thermal cycler (Applied Biosystems Inc., Foster, CA, USA) programmed as follows: 5-min initial denaturation step at 94°C; 35 denaturation cycles at 94°C for 30 s, annealing at 60°C for 1 min and extension at 72°C for 1 min; and final extension at 72°C for 7 min. The annealing temperatures ranged from 55 to 60°C depending on the melting temperature of the primer used.
The amplified DNA fragments and a 10-bp molecular weight marker were separated by 5% denaturing polyacrylamide gel electrophoresis for 90 min. Silver staining was used to visualize the separated fragments according to the method of Bassam et al. (1991)BASSAM, B.J.; CAETANO-ANOLLÉS, G.; GRESSHOFF, P.M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Analytical Biochemistry, v.196, p.80-83, 1991. DOI: 10.1016/0003-2697(91)90120-I.
https://doi.org/10.1016/0003-2697(91)901...
, with modifications. The electrophoretic profiles of the acrylamide gels were photographed and analyzed manually. The presence and absence of alleles for each locus and individual were recorded in an Excel spreadsheet, and statistical analyses were performed.
Genetic diversity was estimated in the nine populations according to the following parameters: Pl, percentage of polymorphic loci in the population; A, average number of alleles; Ae, effective number of alleles (Ae = 1/1 - He) (Hartl & Clark, 1989HARTL, D.L.; CLARK, A.G. Principles of population genetics. 2nded., Sunderland: Sinauer Associates, 1989. 635p.); Ho, observed heterozygosity, calculated from the number of heterozygous individuals divided by the total number of individuals (Nei et al., 1983NEI, M.; TAJIMA, F.; TATENO, Y. Accuracy of estimated phylogenetic trees from molecular data. Journal of Molecular Evolution, v.19, p.153-170, 1983. DOI: 10.1007/BF02300753.
https://doi.org/10.1007/BF02300753....
); and He, expected heterozygosity (He = 1 - Σ Pij ×2, where Pi is the frequency of alleles in the population for each locus). These parameters were calculated using the Popgene software, Version 1.32 (Yeh et al., 1997YEH, F.C.; YANG, R.C.; BOYLE, T.B.J.; YE, Z.H.; MAO, J.X. POPGENE: the user-friendly shareware for population genetic analysis. Edmont: University of Alberta, Molecular Biology and Biotechnology Centre, 1997.). The value of total gene flow (Nm) was estimated from the parameter Fst (Wright, 1949WRIGHT, S. The genetical structure of populations. Annals of Eugenics, v.15, p.323-354, 1949. DOI: 10.1111/j.1469-1809.1949.tb02451.x.
https://doi.org/10.1111/j.1469-1809.1949...
). In addition, paired Nm values were estimated among the following population groups: “wild” populations (P1, P2, and P3), which grew under naturally disturbed conditions or with little impact from human activities and were distant from the crops, paired with the “domestic” populations (P6 and P7), which were planted and cultivated in the traditional milpa system; wild populations (P1, P2, and P3) paired with the “ruderal” populations (P4, P5, and P9), which were grown under conditions disturbed by humans (near roadsides and close to crops); ruderal populations (P4, P5, and P9) paired with the domestic populations (P6 and P7); and population growing spontaneously in a family orchard (P8) paired with the domestic populations (P6 and P7). These parameters were also calculated using the Popgene software, Version 1.32 (Yeh et al., 1997YEH, F.C.; YANG, R.C.; BOYLE, T.B.J.; YE, Z.H.; MAO, J.X. POPGENE: the user-friendly shareware for population genetic analysis. Edmont: University of Alberta, Molecular Biology and Biotechnology Centre, 1997.).
The genetic structure among populations was determined using the independent methods of the Fst estimator (Wright, 1949WRIGHT, S. The genetical structure of populations. Annals of Eugenics, v.15, p.323-354, 1949. DOI: 10.1111/j.1469-1809.1949.tb02451.x.
https://doi.org/10.1111/j.1469-1809.1949...
). The parameters were calculated using the POPGENE software, Version 1.32 (Yeh et al., 1997YEH, F.C.; YANG, R.C.; BOYLE, T.B.J.; YE, Z.H.; MAO, J.X. POPGENE: the user-friendly shareware for population genetic analysis. Edmont: University of Alberta, Molecular Biology and Biotechnology Centre, 1997.). To infer the levels of gene infiltration and probability of ancestry in the studied individuals of the population groups, Bayesian grouping was performed using the Structure software, version 2.3.1 (Pritchard et al., 2000PRITCHARD, J.K.; STEPHENS, M.; DONNELLY, P. Inference of population structure using multilocus genotype data. Genetics, v.155, p.945-959, 2000.). The groups were detected through a combination model that involved the correlation of frequencies between groups (K) and the admixture model for ancestry. In this model, 20 replicates were calculated for K1 to K10 values. The number of clusters (K) that best described the genetic structure of the collection was identified using the Structure Harvester online software, version 0.6.92 (Earl & vonHoldt, 2012EARL, D. A.; VONHOLDT, B.M. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, v.4, p.359-361, 2012. DOI: 10.1007/s12686-011-9548-7.
https://doi.org/10.1007/s12686-011-9548-...
), which adopts the nonparametric statistical method ΔK described by Evanno et al. (2005)EVANNO, G.; REGNAUT, S.; GOUDET, J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology, v.14, p.2611-2620, 2005. DOI: 10.1111/j.1365-294X.2005.02553.x.
https://doi.org/10.1111/j.1365-294X.2005...
with the K2 to K10 values. The replicates of the clustering analyses of the K values were selected and later analyzed using the Clumpp software, version 1.1.2 (Jakobsson & Rosenberg, 2007JAKOBSSON, M.; ROSENBERG, N.A. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics, v.23, p.1801-1806, 2007. DOI: 10.1093/bioinformatics/btm233.
https://doi.org/10.1093/bioinformatics/b...
). The results of the combinations together with the common values of the groups were represented graphically by the Distruct software, version 1.1 (Rosenberg, 2004ROSENBERG, N.A. DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes, v.4, p.137-138, 2004. DOI: 10.1046/j.1471-8286.2003.00566.x.
https://doi.org/10.1046/j.1471-8286.2003...
).
The genealogy of the populations was evaluated by two types of analyses: neighbor-joining using the genetic distance provided by Nei et al. (1983)NEI, M.; TAJIMA, F.; TATENO, Y. Accuracy of estimated phylogenetic trees from molecular data. Journal of Molecular Evolution, v.19, p.153-170, 1983. DOI: 10.1007/BF02300753.
https://doi.org/10.1007/BF02300753....
and non-rooted neighbor-joining tree constructed with the Populations software (Langella, 2002LANGELLA, O. POPULATIONS 1.2.28: population genetic software (individuals or populations distances, phylogenetic trees). 2002. Available at: ˂Available at: ˂http://bioinformatics.org/~tryphon/populations/
˃. Accessed on: Oct. 28 2016.
http://bioinformatics.org/~tryphon/popul...
); and factorial correspondence analysis with allele frequencies obtained by Genetix software, version 4.03 (Belkhir et al., 2004BELKHIR, K.; BORSA, P.; CHIKHI, L.; RAUFASTE, N.; BONHOMME, F. GENETIX 4.05: logiciel sous Windows TM pour la génétique des populations. Montpellier: Université de Montpellier II, Laboratoire Génome, Populations, Interactions, 2004. CNRS UMR 5171.). The allelic displacement was inferred from the values of ancestry (Q) in the highest ΔK. The demographic structure of the population set was composed of the wild and domestic collections. Therefore, it was assumed that within the wild populations, the average Q1 value represented the percentage of wild alleles and the Q2 value represented the percentage of domestic alleles. In the domestic collection, the average Q1 value represented the percentage of wild alleles and the Q2 value represented the percentage of domestic alleles that were not displaced. In this case, the wild ancestry of the wild populations was 1-Q2 domestic, whereas the domesticated ancestry of the domestic populations was 1-Q1 wild. The asymmetry of flow was inferred based on the following calculation: (1-Q wild/1-Q domestic).
Results and Discussion
A total of 87 alleles were detected in the eight pairs of microsatellites used to evaluate C. argyrosperma, compared with the 29 alleles reported for C. pepo and C. moschata (Table 2). The number of alleles per polymorphic SSR locus found in the present study ranged from 5 to 18, with an average of 10.9. The locus with the largest number of alleles observed was CMTm187 (18 alleles), and the loci with the smallest number of alleles were CMTm62 and CMTp131 (five alleles). Ntuli et al. (2015)NTULI, N.R.; TONGOONA, P.B.; ZOBOLO, A.M. Genetic diversity in Cucurbita pepo landraces revealed by RAPD and SSR markers. Scientia Horticulturae, v.189, p.192-200, 2015. DOI: 10.1016/j.scienta.2015.03.020.
https://doi.org/10.1016/j.scienta.2015.0...
, using SSR markers to assess squash (C. pepo), observed a total of 56 alleles, with most loci ranging from 1 to 12 alleles. These authors detected polymorphism in 67.86% of the identified alleles.
The nine populations analyzed showed between 75 and 100% polymorphic loci (Table 3). The number of polymorphic alleles per locus ranged from 1.9 to 4.4. The mean number of alleles per locus was 3.5, indicating that these markers offer good sensitivity for population genetic analyses in C. argyrosperma.Montes-Hernandez & Eguiarte (2002)MONTES-HERNANDEZ, S.; EGUIARTE, L.E. Genetic structure and indirect estimates of gene flow in three taxa of Cucurbita (Cucurbitaceae) in western Mexico. American Journal of Botany, v.89, p.1156-1163, 2002. DOI: 10.3732/ajb.89.7.1156.
https://doi.org/10.3732/ajb.89.7.1156....
evaluated 12 isoenzymatic loci and calculated the genetic diversity of 16 Cucurbita populations from collection sites near those of the present study. These authors found high levels of genetic variation, including a polymorphism rate of 0.96 and a mean allele diversity of 2.08. Cerón González et al. (2010)CERÓN GONZÁLEZ, L.; LEGARIA SOLANO, J.P.; VILLANUEVA VERDUZCO, C.V.; SAHAGÚN CASTELLANOS, J. Diversidad genética en cuatro especies Mexicanas de calabaza (Cucurbita spp.). Revista Fitotecnia Mexicana, v.33, p.189-196, 2010. used RAPD markers to assess populations of C. argyrosperma and identified polymorphisms in 14.77% of loci and 1.14 alleles per locus; these values were lower than those obtained in the present study. The P1 and P2 wild populations showed significantly lower values than the P4 and P8 ruderal populations, though these values were similar to those of the P7 domestic population. The number of effective alleles varied from 1.7 to 3.5. The wild populations were found to have lower values than the weedy population (P4).
The Ho values ranged from 0.24 to 0.38, but the differences were not statistically significant. The He ranged from 0.37 to 0.65 with a mean value of 0.50. The P2 wild population had lower values than the P8 weedy population (Table 3). Montes-Hernandez & Eguiarte (2002)MONTES-HERNANDEZ, S.; EGUIARTE, L.E. Genetic structure and indirect estimates of gene flow in three taxa of Cucurbita (Cucurbitaceae) in western Mexico. American Journal of Botany, v.89, p.1156-1163, 2002. DOI: 10.3732/ajb.89.7.1156.
https://doi.org/10.3732/ajb.89.7.1156....
reported a lower average He of 0.40, but a similar percentage of polymorphic loci when using isoenzime markers (usually less sensitivity in detecting polymorphisms).
Inan et al. (2012)INAN, N.; YILDIZ, M.; SENSOY, S.; KAFKAS, S.; ABAK, K. Efficacy of ISSR and SRAP techniques for molecular characterization of some Cucurbita genotypes including naked (hull-less) seed pumpkin. The Journal of Animal and Plant Sciences, v.22, p.126-136, 2012. found an He value of 0.3 for 24 specimens of Cucurbita. It is possible to conclude that, the differences between the levels of diversity in the present study may be related to the evolutionary scenarios, in which different levels of natural and human-caused selection pressures are observed, and to the effect of agricultural management (including the migration of seeds and fruits).
The genetic diversity of the wild populations was less than or equal to those of the domestic and weedy populations, which may have been due in part to the wild-domestic hybridization, displacement, flood of domesticated alleles from the cultivated populations, or high number of variants that farmers cultivate within a plot, as indicated by Montes-Hernandez & Eguiarte (2002)MONTES-HERNANDEZ, S.; EGUIARTE, L.E. Genetic structure and indirect estimates of gene flow in three taxa of Cucurbita (Cucurbitaceae) in western Mexico. American Journal of Botany, v.89, p.1156-1163, 2002. DOI: 10.3732/ajb.89.7.1156.
https://doi.org/10.3732/ajb.89.7.1156....
. However, the greater genetic diversity in the weedy-domestic populations may have been the product of recombination resulting from the high gene flow promoted by pollinators and human selection in these populations. The recurrent positive selection for individuals with the desired phenotypic traits and the continuous elimination of individuals with undesired wild characteristics must have been an important, but not exclusive, evolutionary force (Zizumbo & Colunga-GarcíaMarín, 2010ZIZUMBO-VILLARREAL, D.; COLUNGA-GARCÍAMARÍN, P. Origin of agriculture and plant domestication in West Mesoamerica. Genetic Resources and Crop Evolution, v.57, p.813-825, 2010. DOI: 10.1007/s10722-009-9521-4.
https://doi.org/10.1007/s10722-009-9521-...
).
Among the wild populations (P1, P2, and P3) growing far from domesticated and domesticated and cultivated populations (P6 and P7), gene flow was determined as Nm = 1.7. Among the wild populations (P1, P2, and P3) and the ruderal populations (P4, P5, and P9) that were close to crops, Nm = 1.9. Among the ruderal populations (P4, P5, and P9), which were close to the crops and the cultivated/domesticated populations, gene flow was Nm = 2.1. Among the weedy population (P8), which grew spontaneously within the crops and the cultivated/domesticated populations (P6 and P7), Nm = 2.7. However, in this case, phenotypic differentiation was low, which indicated that the laxity of human selection on fruits and seeds allowed the genomes of these populations to be domesticated despite their wild-type phenotypes. In a study carried out in Mexico by Cruz-Reyes et al. (2015)CRUZ-REYES, R.; ÁVILA-SAKAR, G.; SÁNCHEZ-MONTOYA, G.; QUESADA, M. Experimental assessment of gene flow between transgenic squash and a wild relative in the center of origin of cucurbits. Ecosphere, v.6, p.1-13, 2015. DOI: 10.1890/ES15-00304.1.
https://doi.org/10.1890/ES15-00304.1....
, the transfer of pollen from domesticated to wild squash (C. argyrosperma subsp. sororia) was evaluated, and gene flow among the populations was shown to generate fertile hybrids.
As the values of gene flow were very high (approximately Nm = 2), a low genetic structure was expected. However, selection of fruits and seeds, particularly from individuals collected or harvested by humans, seems to pose a strong barrier that has allowed genetic differentiation between domestic and wild populations; this phenomenon could be the determining factor in the domestication of the species. In a study by Cerón Gonzáles et al. (2010)CERÓN GONZÁLEZ, L.; LEGARIA SOLANO, J.P.; VILLANUEVA VERDUZCO, C.V.; SAHAGÚN CASTELLANOS, J. Diversidad genética en cuatro especies Mexicanas de calabaza (Cucurbita spp.). Revista Fitotecnia Mexicana, v.33, p.189-196, 2010., the estimated gene flow rate (Nm = 0.14) among Cucurbita spp. was lower than that found in the present study. This lower result indicated that there was less than one migrant per generation, which could also explain the high degree of differentiation between populations. In a previous study, Ntuli et al. (2015)NTULI, N.R.; TONGOONA, P.B.; ZOBOLO, A.M. Genetic diversity in Cucurbita pepo landraces revealed by RAPD and SSR markers. Scientia Horticulturae, v.189, p.192-200, 2015. DOI: 10.1016/j.scienta.2015.03.020.
https://doi.org/10.1016/j.scienta.2015.0...
also found high values of gene flow among squash populations (C. pepo), Nm = 2.23, which indicated a high genetic exchange between them. The main form of gene exchange between the populations of C. pepo resulted from the exchange of seeds between farmers; this effect was also observed in this work and in those of Barboza et al. (2012BARBOZA, N.; ALBERTAZZI, F.J.; SIBAJA-CORDERO, J.A.; MORA-UMAÑA, F.; ASTORGA, C.; RAMÍREZ, P. Analysis of genetic diversity of Cucurbita moschata (D.) germplasm accessions from Mesoamerica revealed by PCR SSCP and chloroplast sequence data. Scientia Horticulturae, v.134, p.60-71, 2012. DOI: 10.1016/j.scienta.2011.10.028.
https://doi.org/10.1016/j.scienta.2011.1...
) and Du et al. (2011)DU, X.; SUN, Y.; LI, X.; ZHOU, J.; LI, X. Genetic divergence among inbred lines in Cucurbita moschata from China. Scientia Horticulturae, v.127, p.207-213, 2011. DOI: 10.1016/j.scienta.2010.10.018.
https://doi.org/10.1016/j.scienta.2010.1...
.
Asymmetric gene flow was found between the wild and domestic populations. Specifically, the gene flow from wild to domestic populations was four times higher than that from domestic to wild populations. This situation can be partially explained by the strong human selection to avoid bitter fruits, an easily distinguishable characteristic of wild populations, in the next generation of seeds. This result indicates that the asymmetry of the flow is approximately three times greater than that reported in beans by Papa & Gepts (2003)PAPA, R.; GEPTS, P. Asymmetry of gene flow and differential geographical structure of molecular diversity in wild and domesticated common bean (Phaseolus vulgaris L.) from Mesoamerica. Theoretical and Applied Genetics, v.106, p.239-250, 2003. DOI: 10.1007/s00122-002-1085-z.
https://doi.org/10.1007/s00122-002-1085-...
and Martínez-Salvador et al. (2007)MARTÍNEZ-SALVADOR, M.; BELTRÁN-MORALES, L.; VALDEZ-CEPEDA, R.; RUBIO ARIAS, H.; TROYO-DIEGUEZ, E.; MURILLO-AMADOR, B.; JIMÉNEZ GALINDO, J.; ORTEGA-RUBIO, A. Assessment of sustainability performance on the utilization of Agave (Agave salmiana ssp. crassispina) in Zacatecas, México. International Journal of Sustainable Development and World Ecology, v.14, p.362-371, 2007. DOI: 10.1080/13504500709469736.
https://doi.org/10.1080/1350450070946973...
. Moreover, this result can be explained in part by the greater efficiency of cross-breeding and more strict selection in Cucurbita.
The two estimators of genetic structure - Fst and analysis of molecular variance (Amova) - were high in the studied populations. A value of Fst = 0.31 was obtained, indicating that 31% of the diversity occured between populations and 69% within populations, while the Amova results indicated that 31.8% of the diversity was observed between populations and 68.2% within populations. These results indicate high genetic differentiation between populations despite their high gene flow. Genetic differentiation within populations can be explained by the genetic distance between wild and domesticated populations, which is a product of generational seed selection by farmers and the result of natural selection in different environments. It is important to note that, currently, isolated wild populations growing under natural vegetation are rapidly disappearing due to habitat destruction, expansion of agriculture, and influences of livestock and florists. Furthermore, these populations are located in places that are practically inaccessible, making collection very difficult (Zizumbo-Vilarreal & Colunga-GarcíaMarín, 2010ZIZUMBO-VILLARREAL, D.; COLUNGA-GARCÍAMARÍN, P. Origin of agriculture and plant domestication in West Mesoamerica. Genetic Resources and Crop Evolution, v.57, p.813-825, 2010. DOI: 10.1007/s10722-009-9521-4.
https://doi.org/10.1007/s10722-009-9521-...
).
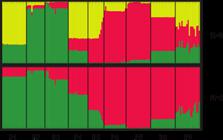
The K-estimator test indicated that the highest values of K, which best describe the demographic structure between K1 and K10, were K = 2 (5.3) and K = 3 (2.7) (Figure 1). When the best value of K = 2 was obtained, the demographic structure of the global stock was composed of populations with Q1 values ranging from 85 to 99%. The wild populations that grew spontaneously under conditions disturbed naturally or by human activity and far from crops (P1, P2, and P3) and those populations that grew spontaneously or were cultivated in agricultural sites (P6, P7, and P8) presented Q2 values between 80 and 85% (Figure 1). The next highest value indicated that P1, a population that grew under natural conditions far from the crops, was not isolated. However, at K = 3, the demographic structure suggested that the genetic differentiation of the three types of populations was determined by natural selective pressures in the areas disturbed by human activity outside the plots. At K = 5, the demographic structure suggested that the specificity of human disturbance and selective pressures during consumption were the most important factors.
Stratified demographic structure for K = 2 and K = 3 values considering the nine squash (Cucurbita argyrosperma) populations (P1 to P9) studied with simple sequence repeat markers. Each color represents a different biological-evolutionary state in the evaluated populations.
At K = 3, the first collection consisted of populations with Q values greater than 80% (P2 and P3), whereas the second collection was formed by the P7 domesticated population. The third collection was a mixture in which P1 had the highest values of Q3 (70%). Considering that the set of populations studied includes two wild collections, the mean value of Q1 in the wild populations (P1 and P2) was 0.87%, while Q2 was 0.13. In other words, the wild populations consisted of, on average, 87% wild alleles and 13% domestic alleles, whereas the domestic populations (P6 and P7) presented, on average, values of Q1 = 0.3 and Q2 = 0.97. This result can be explained by the fact that the domestic populations are made up of 97% domestic alleles and 3% wild alleles. That is, domestic allelic displacement in the wild collection accounts for 13%, and wild displacement in the domestic collection represents 3%. Therefore, there has been asymmetry in gene flow, with domesticated genes flowing into wild populations at a rate 4.3 times that of wild genes flowing into domesticated populations. Considering the next highest value of K = 3, the studied population consisted of the wild (P2 and P3), the domestic (P6 and P7), and the ruderal or weedy (P4, P5, and P9) collections. However, under this condition, none of the collections presented Q3 values greater than 85% of the weedy collection. The greatest genetic distance between the evaluated populations was between the P2 wild population and the P5 ruderal population (0.88) (Table 4). The populations with the lowest genetic distance were the P2 and P3 wild populations (0.41).
Genetic distance values analyzed with simple sequence repeat markers among the nine squash (Cucurbita argyrosperma) populations (P1 to P9) from the states of Jalisco and Colima, Mexico.
The weedy populations showed high values of wild genome displacement, considering 78.5% of their genome was domesticated. This result can be explained in part by the migration of pollen and seeds from domestic plants, since the traditional harvesting of domestic, weedy, and domestic-weedy hybrid seeds occurs in the plot and the seeds of all the fruits remain on the ground until the end of the growing season; these remaining seeds eventually germinate, creating back-crosses that grow and are tolerated in the plot. In populations growing in disturbed areas relatively close to or around crops, the displacement of alleles by domesticated ones was 23.5% in the wild genome. This result can be partly attributed to the high migration of pollen from crops and the adaptive capacity of hybrids and back-crosses under conditions of human disturbance. Reproductive isolation through the selection of mutants with short lifespans and cultivation outside the distribution of wild individuals (geographic isolation) has played a very important role. In addition to new mutations, genetic expression and environmental conditions induced by the crop affect new phenotypes and the origins of genetic pools with high phenotypic diversity; these plants are subsequently collected, selected, and disseminated by humans over a long period of time (Lira et al., 2016LIRA, R.; EGUIARTE L.; MONTES, S.; ZIZUMBO-VILLARREAL, D.; COLUNGA-GARCÍAMARIN, P.; QUESADA, M. Homo sapiens-Cucurbita interaction in Mesoamerica: domestication, dissemination, and diversification. In: LIRA, R.; CASAS, A.; BLANCAS, J. (Ed.). Ethnobotany of Mexico: interactions of people and plants in Mesoamerica. New York: Springer, 2016. p.389-400. DOI: 10.1007/978-1-4614-6669-7_15.
https://doi.org/10.1007/978-1-4614-6669-...
).
Despite the high genetic displacement of the wild genome by the domestic one due to the reproductive biology of the species, the sympatry of domestic and wild populations, and the presence of pollinators, only 3% of the domestic genome was displaced by the wild one, indicating the importance of human selective pressure. This type of pressure operates at the same level as geographic isolation, as 97% of the wild genome remained in the population isolated from the crops (Q = 0.97); i.e., only 3% of the the wild genome was displaced. Traditional squash farmers have recognized and maintained diversity for many years primarily by continuously cultivating empirically selected segregants, which appear spontaneously in the local varieties. In each crop cycle, farmers select the best fruits based on certain characteristics (such as diameter, shape, and external texture of the fruit; color and flavor of the pulp; and length, weight, and width of fruits and seeds) to obtain seeds for propagation in the next cycle. They also select plants taking into account growth characteristics, flowering patterns, and resistance to pests, fungi, viruses, and weeds (Ferriol & Picó, 2008FERRIOL, M.; PICÓ, B. Pumpkin and winter squash. In: PROHENS, J.; NUEZ, F. (Ed.). Vegetables I:(Asteraceae, Brassicaceae, Chenopodicaceae, and Cucurbitaceae. New York: Springer, 2008. p.317-349. (Handbook of Plant Breeding). DOI: 10.1007/978-0-387-30443-4_10.
https://doi.org/10.1007/978-0-387-30443-...
; Sánchez-Hernández et al., 2014SÁNCHEZ-HERNÁNDEZ, M.Á.; VILLANUEVA-VERDUZCO, C.; SÁNCHEZ-HERNÁNDEZ, C.; SAHAGÚN-CASTELLANOS, J.; VILLANUEVA-SÁNCHEZ, E. Respuesta a la selección participativa en variedades de calabaza de la Sierra Norte de Puebla, México, Revista Chapingo Serie Horticultura, v.20, p.41-56, 2014 .DOI: 10.5154/r.rchsh.2011.10.058.
https://doi.org/10.5154/r.rchsh.2011.10....
).
The results of clustering using the neighbor-joining analysis indicated three genealogically related groups that grow in different agroecological areas (Figure 2). Group I consisted of populations of C. argyrosperma subsp. sororia that were eventually harvested and consumed, or P1, P2, and P3, which grew far from the crops and under natural or continuously disturbed conditions. Group II was formed by individuals from the P6 and P7 domesticated populations of C. argyrosperma subsp. argyrosperma, as well as from P8, which grew spontaneously in the harvested and consumed plots. Group III consisted of the P4, P5, and P9 ruderal populations, which grew close to the crops under disturbed conditions and were eventually harvested and consumed. A similar phenomenon was observed in the study by Ntuli et al. (2015)NTULI, N.R.; TONGOONA, P.B.; ZOBOLO, A.M. Genetic diversity in Cucurbita pepo landraces revealed by RAPD and SSR markers. Scientia Horticulturae, v.189, p.192-200, 2015. DOI: 10.1016/j.scienta.2015.03.020.
https://doi.org/10.1016/j.scienta.2015.0...
, in which C. pepo populations were grouped according to the agroecological regions where they were cultivated.
Dendrogram of the nine studied populations (P1 to P9) of squash (Cucurbita argyrosperma) obtained by non-rooted neighbor-joining clustering analysis based on the genetic distance matrix provided by Nei et al. (1983)NEI, M.; TAJIMA, F.; TATENO, Y. Accuracy of estimated phylogenetic trees from molecular data. Journal of Molecular Evolution, v.19, p.153-170, 1983. DOI: 10.1007/BF02300753.
https://doi.org/10.1007/BF02300753.... . Sequences were amplified using eight simple sequence repeat markers. The values found in the groups indicate the percentage of times the populations were grouped together with 1,000 bootstrapping cycles.
The ethnobotanical analyses indicated that both the fruits and seeds of wild and domesticated populations are harvested and consumed by local human populations. The fruits of the wild and weedy populations can be consumed after being washed with oven ashes, for example; this possibly archaic method favors human interest and could have led to domestication (Zizumbo-Villarreal et al., 2012ZIZUMBO-VILLARREAL, D.; FLORES-SILVA, A.; COLUNGA-GARCÍA MARÍN, P. The Archaic diet in Mesoamerica: incentive for milpa development and species domestication. Economic Botany, v.66, p.328-343, 2012. DOI: 10.1007/s12231-012-9212-5.
https://doi.org/10.1007/s12231-012-9212-...
). According to these authors, the seeds of domestic populations, as well as those consumed by producers, are marketed, representing an important economic resource for local families and indicating that both wild and domesticated populations are subject to human-based selection pressures at different scales (Zizumbo-Villarreal et al., 2014ZIZUMBO-VILLARREAL, D.; FLORES-SILVA, A.; COLUNGA-GARCÍAMARÍN, P. The food system during the formative period in west Mesoamerica. Economic Botany, v.68, p.67-84, 2014. DOI: 10.1007/s12231-014-9262-y.
https://doi.org/10.1007/s12231-014-9262-...
).
Besides the neighboring-joining analysis, the factorial correspondence analysis was also used. It pointed out the same three population groups, explaining 52% of the genetic variation in the three obtained axes (Figure 3).
Factorial correspondence analysis for the nine populations (P1 to P9) of squash (Cucurbita argyrosperma) studied with simple sequence repeat markers.
It should be noted that the abovementioned studies are of great importance for designing biosafety strategies, considering the degree to which human or natural selection affects populations. With the observed results, it is possible to plan biosecurity strategies for managing these food resources, as well as conservation efforts for this species.
Conclusions
-
The current genetic heritage of wild-weedy-domestic populations of squash (Cucurbita argyrosperma) is determined in five different ecological scenarios, under different pressures of natural and human-based selection.
-
There is high gene flow in the wild-weedy-domestic populations of squash.
-
The greatest genetic diversity is found in weedy-domestic populations.
Acknowledgments
To Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Capes); to Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq); and to Centro de Investigación Científica de Yucatán (CICY), for technical support.
References
- BARBOZA, N.; ALBERTAZZI, F.J.; SIBAJA-CORDERO, J.A.; MORA-UMAÑA, F.; ASTORGA, C.; RAMÍREZ, P. Analysis of genetic diversity of Cucurbita moschata (D.) germplasm accessions from Mesoamerica revealed by PCR SSCP and chloroplast sequence data. Scientia Horticulturae, v.134, p.60-71, 2012. DOI: 10.1016/j.scienta.2011.10.028.
» https://doi.org/10.1016/j.scienta.2011.10.028. - BASSAM, B.J.; CAETANO-ANOLLÉS, G.; GRESSHOFF, P.M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Analytical Biochemistry, v.196, p.80-83, 1991. DOI: 10.1016/0003-2697(91)90120-I.
» https://doi.org/10.1016/0003-2697(91)90120-I. - BELKHIR, K.; BORSA, P.; CHIKHI, L.; RAUFASTE, N.; BONHOMME, F. GENETIX 4.05: logiciel sous Windows TM pour la génétique des populations. Montpellier: Université de Montpellier II, Laboratoire Génome, Populations, Interactions, 2004. CNRS UMR 5171.
- CERÓN GONZÁLEZ, L.; LEGARIA SOLANO, J.P.; VILLANUEVA VERDUZCO, C.V.; SAHAGÚN CASTELLANOS, J. Diversidad genética en cuatro especies Mexicanas de calabaza (Cucurbita spp.). Revista Fitotecnia Mexicana, v.33, p.189-196, 2010.
- CRUZ-REYES, R.; ÁVILA-SAKAR, G.; SÁNCHEZ-MONTOYA, G.; QUESADA, M. Experimental assessment of gene flow between transgenic squash and a wild relative in the center of origin of cucurbits. Ecosphere, v.6, p.1-13, 2015. DOI: 10.1890/ES15-00304.1.
» https://doi.org/10.1890/ES15-00304.1. - DU, X.; SUN, Y.; LI, X.; ZHOU, J.; LI, X. Genetic divergence among inbred lines in Cucurbita moschata from China. Scientia Horticulturae, v.127, p.207-213, 2011. DOI: 10.1016/j.scienta.2010.10.018.
» https://doi.org/10.1016/j.scienta.2010.10.018. - EARL, D. A.; VONHOLDT, B.M. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, v.4, p.359-361, 2012. DOI: 10.1007/s12686-011-9548-7.
» https://doi.org/10.1007/s12686-011-9548-7. - ECHEVARRÍA-MACHADO, I.; SÁNCHEZ-CACH, L.A.; HERNÁNDEZ-ZEPEDA, C.; RIVERA-MADRID, R.; MORENO-VALENZUELA, O.A. A simple and efficient method for isolation of DNA in high mucilaginous plant tissues. Molecular Biotechnology, v.31, p.129-135, 2005. DOI: 10.1385/MB:31:2:129.
» https://doi.org/10.1385/MB:31:2:129. - EVANNO, G.; REGNAUT, S.; GOUDET, J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology, v.14, p.2611-2620, 2005. DOI: 10.1111/j.1365-294X.2005.02553.x.
» https://doi.org/10.1111/j.1365-294X.2005.02553.x. - FERRIOL, M.; PICÓ, B. Pumpkin and winter squash. In: PROHENS, J.; NUEZ, F. (Ed.). Vegetables I:(Asteraceae, Brassicaceae, Chenopodicaceae, and Cucurbitaceae. New York: Springer, 2008. p.317-349. (Handbook of Plant Breeding). DOI: 10.1007/978-0-387-30443-4_10.
» https://doi.org/10.1007/978-0-387-30443-4_10. - HARTL, D.L.; CLARK, A.G. Principles of population genetics. 2nded., Sunderland: Sinauer Associates, 1989. 635p.
- HUFFORD, M.B.; LUBINKSY, P.; PYHÄJÄRVI, T.; DEVENGENZO, M.T.; ELLSTRAND, N.C.; ROSS-IBARRA, J. The genomic signature of crop-wild introgression in maize. Plos Genetics, v.9, e1003477, 2013. DOI: 10.1371/annotation/2eef7b5b-29b2-412f-8472-8fd7f9bd65ab.
» https://doi.org/10.1371/annotation/2eef7b5b-29b2-412f-8472-8fd7f9bd65ab. - INAN, N.; YILDIZ, M.; SENSOY, S.; KAFKAS, S.; ABAK, K. Efficacy of ISSR and SRAP techniques for molecular characterization of some Cucurbita genotypes including naked (hull-less) seed pumpkin. The Journal of Animal and Plant Sciences, v.22, p.126-136, 2012.
- JAKOBSSON, M.; ROSENBERG, N.A. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics, v.23, p.1801-1806, 2007. DOI: 10.1093/bioinformatics/btm233.
» https://doi.org/10.1093/bioinformatics/btm233. - LANGELLA, O. POPULATIONS 1.2.28: population genetic software (individuals or populations distances, phylogenetic trees). 2002. Available at: ˂Available at: ˂http://bioinformatics.org/~tryphon/populations/ ˃. Accessed on: Oct. 28 2016.
» http://bioinformatics.org/~tryphon/populations/ - LIRA SAADE, R. Estudios taxonómicos y ecogeográficos de las cucurbitaceae latinoamericanas de importancia económica: Cucurbita, Sechium, Sicana y Cyclanthera Rome: International Plant Genetic Resources Institute, 1995. 281p. (Systematic and Ecogeographic Studies on Crop Gene Pools, v. 9).
- LIRA, R.; EGUIARTE L.; MONTES, S.; ZIZUMBO-VILLARREAL, D.; COLUNGA-GARCÍAMARIN, P.; QUESADA, M. Homo sapiens-Cucurbita interaction in Mesoamerica: domestication, dissemination, and diversification. In: LIRA, R.; CASAS, A.; BLANCAS, J. (Ed.). Ethnobotany of Mexico: interactions of people and plants in Mesoamerica. New York: Springer, 2016. p.389-400. DOI: 10.1007/978-1-4614-6669-7_15.
» https://doi.org/10.1007/978-1-4614-6669-7_15. - MARTÍNEZ-SALVADOR, M.; BELTRÁN-MORALES, L.; VALDEZ-CEPEDA, R.; RUBIO ARIAS, H.; TROYO-DIEGUEZ, E.; MURILLO-AMADOR, B.; JIMÉNEZ GALINDO, J.; ORTEGA-RUBIO, A. Assessment of sustainability performance on the utilization of Agave (Agave salmiana ssp. crassispina) in Zacatecas, México. International Journal of Sustainable Development and World Ecology, v.14, p.362-371, 2007. DOI: 10.1080/13504500709469736.
» https://doi.org/10.1080/13504500709469736. - MERRICK, L.C. Systematics and evolution of a domesticated squash, Cucurbita argyrosperma, and its wild and weedy relatives. In: BATES, D.M.; ROBINSON, R.W.; JEFFREY, C. (Ed.). Biology and utilization of the Cucurbitaceae. Ithaca: Cornell University Press, 1990. p.77-95.
- MONTES-HERNANDEZ, S.; EGUIARTE, L.E. Genetic structure and indirect estimates of gene flow in three taxa of Cucurbita (Cucurbitaceae) in western Mexico. American Journal of Botany, v.89, p.1156-1163, 2002. DOI: 10.3732/ajb.89.7.1156.
» https://doi.org/10.3732/ajb.89.7.1156. - NEI, M.; TAJIMA, F.; TATENO, Y. Accuracy of estimated phylogenetic trees from molecular data. Journal of Molecular Evolution, v.19, p.153-170, 1983. DOI: 10.1007/BF02300753.
» https://doi.org/10.1007/BF02300753. - NTULI, N.R.; TONGOONA, P.B.; ZOBOLO, A.M. Genetic diversity in Cucurbita pepo landraces revealed by RAPD and SSR markers. Scientia Horticulturae, v.189, p.192-200, 2015. DOI: 10.1016/j.scienta.2015.03.020.
» https://doi.org/10.1016/j.scienta.2015.03.020. - PAPA, R.; GEPTS, P. Asymmetry of gene flow and differential geographical structure of molecular diversity in wild and domesticated common bean (Phaseolus vulgaris L.) from Mesoamerica. Theoretical and Applied Genetics, v.106, p.239-250, 2003. DOI: 10.1007/s00122-002-1085-z.
» https://doi.org/10.1007/s00122-002-1085-z. - PAYRÓ DE LA CRUZ, E.; GEPTS, P.; COLUNGA GARCIAMARÍN, P.; ZIZUMBO VILLARREAL, D. Spatial distribution of genetic diversity in wild populations of Phaseolus vulgaris L. from Guanajuato and Michoacán, México. Genetic Resources and Crop Evolution, v.52, p.589-599, 2005. DOI: 10.1007/s10722-004-6125-x.
» https://doi.org/10.1007/s10722-004-6125-x. - PIÑEYRO-NELSON, A.; VAN HEERWAARDEN, J.; PERALES, H.R.; SERRATOS- HERNÁNDEZ, J.A.; RANGEL, A.; HUFFORD, M.B.; GEPTS, P.; GARAY-ARROYO, A.; RIVERA-BUSTAMANTE, R.; ÁLVAREZ-BUYLLA, E.R. Transgenes in Mexican maize: molecular evidence and methodological considerations for GMO detection in landrace populations. Molecular Ecology, v.18, p.750-761, 2009. DOI: 10.1111/j.1365-294X.2008.03993.x.
» https://doi.org/10.1111/j.1365-294X.2008.03993.x. - PRITCHARD, J.K.; STEPHENS, M.; DONNELLY, P. Inference of population structure using multilocus genotype data. Genetics, v.155, p.945-959, 2000.
- ROSENBERG, N.A. DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes, v.4, p.137-138, 2004. DOI: 10.1046/j.1471-8286.2003.00566.x.
» https://doi.org/10.1046/j.1471-8286.2003.00566.x. - SÁNCHEZ-HERNÁNDEZ, M.Á.; VILLANUEVA-VERDUZCO, C.; SÁNCHEZ-HERNÁNDEZ, C.; SAHAGÚN-CASTELLANOS, J.; VILLANUEVA-SÁNCHEZ, E. Respuesta a la selección participativa en variedades de calabaza de la Sierra Norte de Puebla, México, Revista Chapingo Serie Horticultura, v.20, p.41-56, 2014 .DOI: 10.5154/r.rchsh.2011.10.058.
» https://doi.org/10.5154/r.rchsh.2011.10.058. - SANJUR, O.I.; PIPERNO, D.R.; ANDRES, T.C.; WESSEL-BEAVER, L. Phylogenetic relationships among domesticated and wild species of Cucurbita (Cucurbitaceae) inferred from a mitochondrial gene: implications for crop plant evolution and areas of origin. Proceedings of the National Academy Sciences of the United States of America, v.99, p.535-540, 2002. DOI: 10.1073/pnas.012577299.
» https://doi.org/10.1073/pnas.012577299. - WILSON, H.D.; LIRA, R.; RODRÍGUEZ, I. Crop/weed gene flow: Cucurbita argyrosperma Huber and C. fraterna L.H. Bailey (Cucurbitaceae). Economic Botany, v.48, p.293-300, 1994. DOI: 10.1007/BF02862330.
» https://doi.org/10.1007/BF02862330. - WRIGHT, S. The genetical structure of populations. Annals of Eugenics, v.15, p.323-354, 1949. DOI: 10.1111/j.1469-1809.1949.tb02451.x.
» https://doi.org/10.1111/j.1469-1809.1949.tb02451.x. - YEH, F.C.; YANG, R.C.; BOYLE, T.B.J.; YE, Z.H.; MAO, J.X. POPGENE: the user-friendly shareware for population genetic analysis. Edmont: University of Alberta, Molecular Biology and Biotechnology Centre, 1997.
- ZIZUMBO-VILLARREAL, D.; COLUNGA-GARCÍAMARÍN, P. Origin of agriculture and plant domestication in West Mesoamerica. Genetic Resources and Crop Evolution, v.57, p.813-825, 2010. DOI: 10.1007/s10722-009-9521-4.
» https://doi.org/10.1007/s10722-009-9521-4. - ZIZUMBO-VILLARREAL, D.; FLORES-SILVA, A.; COLUNGA-GARCÍA MARÍN, P. The Archaic diet in Mesoamerica: incentive for milpa development and species domestication. Economic Botany, v.66, p.328-343, 2012. DOI: 10.1007/s12231-012-9212-5.
» https://doi.org/10.1007/s12231-012-9212-5. - ZIZUMBO-VILLARREAL, D.; FLORES-SILVA, A.; COLUNGA-GARCÍAMARÍN, P. The food system during the formative period in west Mesoamerica. Economic Botany, v.68, p.67-84, 2014. DOI: 10.1007/s12231-014-9262-y.
» https://doi.org/10.1007/s12231-014-9262-y. - ZIZUMBO-VILLARREAL, D.; GONZÁLEZ-ZOZAYA, F.; OLAY-BARRIENTOS, A.; PLATAS-RUÍZ, R.; CUEVAS-SAGARDÍ, M.; ALMENDROS-LÓPEZ, L.; COLUNGA-GARCÍAMARÍN, P. Archaeological evidence of the cultural importance of Agave spp in pre-hispanic Colima, Mexico. Economic Botany, v.63, p.288-302, 2009.
Publication Dates
-
Publication in this collection
Mar 2018
History
-
Received
28 Oct 2016 -
Accepted
28 June 2017