Abstracts
PURPOSE: This study aimed to evaluate the frequency of homozygous deletion of GSTM1 and GSTT1 genes and their combinations between patients with breast cancer and healthy individuals, associating them with disease susceptibility. METHODS: This is a case-control study in which 49 women diagnosed with breast cancer confirmed by pathological examination and 49 healthy women with no evidence of cancer and no prior family history of breast cancer were invited to participate. All of them answered a questionnaire with epidemiological data and were submitted to blood sample collection. Genomic DNA was extracted from blood, and genotyping was performed by polymerase chain reaction. Data were analyzed with SPSS 20.0. RESULTS: The frequency of null alleles for GSTM1 and GSTT1 was 58.8 and 61.7%, respectively, for patients with breast cancer, and 41.2 and 38.3%, respectively, in control patients. In homozygous deletion of the GSTM1 gene, a significantly higher frequency was found in the breast cancer cases. CONCLUSION: Breast cancer patients presented higher frequency of homozygous deletion of the GSTM1 gene compared with the control group.
Breast neoplasms; Breast neoplasms; Genetic predisposition to disease; Glutathione transferase; Disease susceptibility; Case-control studies
OBJETIVO: Este estudo teve como objetivo avaliar a frequência de deleção homozigótica dos genes GSTM1 e GSTT1 e suas combinações entre os pacientes com câncer de mama e indivíduos saudáveis, associando-se a suscetibilidade à doença. MÉTODOS: Este é um estudo de caso-controle, no qual 49 mulheres diagnosticadas com câncer de mama confirmado por exame anatomopatológico e 49 mulheres saudáveis, sem evidência de câncer e sem história familiar prévia de câncer de mama, foram convidadas a participar. Todas responderam a um questionário com dados epidemiológicos e foram submetidas à coleta de sangue. O DNA foi extraído a partir de sangue, e genotipagem foi realizada por reação em cadeia da polimerase. Os dados foram analisados com o SPSS 20.0. RESULTADOS: A frequência de alelos nulos para GSTM1 e GSTT1 foi de 58,8 e 61,7%, respectivamente, para as pacientes com câncer de mama, e 41,2 e 38,3%, respectivamente, em pacientes do grupo controle. Em deleção homozigótica do gene GSTM1, uma frequência significativamente maior foi encontrada nos casos. CONCLUSÃO: Pacientes com câncer de mama apresentam uma maior frequência de deleção homozigótica do gene GSTM1 quando comparadas com o grupo controle.
Neoplasias da mama; Neoplasias da mama; Predisposição genética para doença; Glutationa transferase; Susceptibilidade a doenças; Estudos de casos e controles
ORIGINAL ARTICLE
Polymorphisms of GSTM1 and GSTT1 genes in breast cancer susceptibility: a case-control study
Polimorfismo dos genes GSTM1 e GSTT1 na suscetibilidade do câncer de mama: estudo caso-controle
Lia Gonçalves PossueloI; Camila Farias PeraçaII; Michelle Fraga EisenhardtII; Marcelo Luis DottoIII; Lucas CappellettiIV; Eliara FolettoII; Andreia Rosane de Moura ValimIV
IPostgraduate Program in Health Promotion, Universidade de Santa Cruz do Sul - UNISC - Santa Cruz do Sul (RS), Brazil
IIPharmacy Graduation Course, Universidade de Santa Cruz do Sul - UNISC - Santa Cruz do Sul (RS), Brazil
IIIIntegrated Oncology Center, Hospital Ana Nery - Santa Cruz do Sul (RS), Brazil
IVMedicine Graduation Course, Universidade de Santa Cruz do Sul - UNISC - Santa Cruz do Sul (RS), Brazil
Correspondence Correspondence: Lia Gonçalves Possuelo Av. Independência, 2.293, Bloco 35, sala 3.504 CEP: 96815-900 Santa Cruz do Sul (RS), Brazil
ABSTRACT
PURPOSE: This study aimed to evaluate the frequency of homozygous deletion of GSTM1 and GSTT1 genes and their combinations between patients with breast cancer and healthy individuals, associating them with disease susceptibility.
METHODS: This is a case-control study in which 49 women diagnosed with breast cancer confirmed by pathological examination and 49 healthy women with no evidence of cancer and no prior family history of breast cancer were invited to participate. All of them answered a questionnaire with epidemiological data and were submitted to blood sample collection. Genomic DNA was extracted from blood, and genotyping was performed by polymerase chain reaction. Data were analyzed with SPSS 20.0.
RESULTS: The frequency of null alleles for GSTM1 and GSTT1 was 58.8 and 61.7%, respectively, for patients with breast cancer, and 41.2 and 38.3%, respectively, in control patients. In homozygous deletion of the GSTM1 gene, a significantly higher frequency was found in the breast cancer cases.
CONCLUSION: Breast cancer patients presented higher frequency of homozygous deletion of the GSTM1 gene compared with the control group.
Keywords: Breast neoplasms/genetics; Breast neoplasms/enzimology; Genetic predisposition to disease; Glutathione transferase/genetics; Disease susceptibility; Case-control studies.
RESUMO
OBJETIVO: Este estudo teve como objetivo avaliar a frequência de deleção homozigótica dos genes GSTM1 e GSTT1 e suas combinações entre os pacientes com câncer de mama e indivíduos saudáveis, associando-se a suscetibilidade à doença.
MÉTODOS: Este é um estudo de caso-controle, no qual 49 mulheres diagnosticadas com câncer de mama confirmado por exame anatomopatológico e 49 mulheres saudáveis, sem evidência de câncer e sem história familiar prévia de câncer de mama, foram convidadas a participar. Todas responderam a um questionário com dados epidemiológicos e foram submetidas à coleta de sangue. O DNA foi extraído a partir de sangue, e genotipagem foi realizada por reação em cadeia da polimerase. Os dados foram analisados com o SPSS 20.0.
RESULTADOS: A frequência de alelos nulos para GSTM1 e GSTT1 foi de 58,8 e 61,7%, respectivamente, para as pacientes com câncer de mama, e 41,2 e 38,3%, respectivamente, em pacientes do grupo controle. Em deleção homozigótica do gene GSTM1, uma frequência significativamente maior foi encontrada nos casos.
CONCLUSÃO: Pacientes com câncer de mama apresentam uma maior frequência de deleção homozigótica do gene GSTM1 quando comparadas com o grupo controle.
Palavras-chave: Neoplasias da mama/genética; Neoplasias da mama/enzimologia; Predisposição genética para doença; Glutationa transferase/genética; Susceptibilidade a doenças; Estudos de casos e controles.
Introduction
Breast cancer is now the most common cancer both in developed and developing regions with 690,000 new cases estimated in each region (population ratio 1:4). Incidence rates vary from 19.3 per 100,000 women in Eastern Africa to 89.9 per 100,000 women in Western Europe, and are high (greater than 80 per 100,000) in developed regions of the world (except Japan)1.
A total of 49,240 new cases of breast cancer were estimated for Brazil in 2011, and Rio Grande do Sul is one of the states with the highest estimate in the country, with 81.57 cases per 100,000 women2.
However, breast cancer can also be considered to be sporadic, representing more than 90% of cases worldwide. Clinical, epidemiological and experimental studies have shown that the risk of developing sporadic breast cancer is related to the production of sex steroids. Thus, the endocrine conditions modulated by ovarian function, such as early menarche, late menopause and pregnancy, as well as the use of exogenous estrogens, are important components of the risk of developing breast cancer3.
In recent years, there have been great advances in the knowledge of genetic polymorphisms that affect both the enzymes involved in activation and detoxification of certain chemotherapeutic agents as well as molecular targets involved in cancer treatment. A correct identification of patients with the hereditary forms of breast cancer and a molecular approach for detection of associated mutations are still a challenge in the genetic context worldwide. Thus, there is a constant interest to study gene polymorphisms that may be associated with the disease4.
The Glutathione S-transferases (GSTs) are a family of Phase II intracellular enzymes that catalyze cell detoxification, protecting them from attack by reactive electrophiles harmful to cells, also known as xenobiotics, thus preventing possible damage to cellular DNA. This detoxification occurs through a combination of glutathione (GSH) with a wide variety of endogenous and exogenous electrophilic compounds5,6. Therefore, it is possible to suppose that the lack of these genes may increase the risk of developing neoplasia7,8.
The GSTT1 and GSTM1 genes, which belong to the family of GSTs, encode the major proteins involved in the conjugation of the substrates that are toxic to cells. Both genes are highly polymorphic and can be present or deleted in homozygous form9,10. Women with deletion of these genes have a three-fold higher risk of developing breast cancer11.
The GSTT1 (AB057594) gene is located on chromosome 22q11.23, has 8,146 base pairs and consists of 5 exons and 4 introns that encode a protein with 240 amino acids. This gene has important differences in its catalytic activity when compared with others in the GST family and is considered one of the oldest members of GST family10. The GSTM1 gene (AY532926) is located on chromosome 1p13.3 and has 657 base pairs. Individuals with the homozygous null genotype for GSTM1 and GSTT1 do not express these proteins. Thus, the absence of these genes can result in an increased accumulation of reactive metabolites in the body, increasing the likelihood of interaction with cellular macromolecules and initiate the tumorigenesis process12.
However, this study aimed to assess the frequency of homozygous deletions of GSTT1 and GSTM1 genes in women with breast cancer and in women without breast cancer, and to verify whether the occurrence of deletions is associated with clinical factors, sporadic or familial breast cancer and with increased susceptibility to breast cancer development.
Methods
Study design and patients
We conducted a case-control study in which the population consisted of 49 women diagnosed with breast cancer (cases), confirmed by pathological examination, with or without immunohistochemistry, treated at the Center for Integrative Oncology (COI) of Hospital Ana Nery, and 49 healthy women (controls) with no evidence of cancer and no prior family history of breast cancer seen at the Integrated Health Service (SIS) at University of Santa Cruz do Sul (UNISC) during gynecological examinations routine, in Santa Cruz do Sul (RS), Brazil. The sample was included by convenience. Data collection occurred from March to August 2011. Both groups were matched for age. The women were invited to participate in the study and agreed by signing the Free and Informed Consent Form (FICF). This work is in accordance with resolution 196/96 and was approved by the Ethics Committee (CEP-UNISC) under protocol number 2730/2010.
Both groups, cases and controls, answered an epidemiological questionnaire with information related to smoking, oral contraceptive use, use of hormone replacement therapy in postmenopausal women, age (years), age at menarche, age at menopause, age at first pregnancy, duration of breastfeeding, and history of another cancer in the family; additionally, the case group had their medical files reviewed to extract information such as age of diagnosis of breast cancer, disease stage, type of treatment received (chemotherapy, radiotherapy and/or hormone therapy) and therapeutic regimen, when they received chemotherapy.
Regarding the presence of family history of breast cancer, all women who reported a family history of either first or second-degree relatives formed a sub-group of familial breast cancer cases. The others, who reported no relatives with the disease, were included in the sub-group of sporadic cases of breast cancer. Both sub-groups belonged to the case group.
Molecular analysis
All women underwent a peripheral blood sample collection for genetic analysis in the Laboratory of Genetics and Biotechnology of UNISC. Genomic DNA was extracted from peripheral blood leukocytes by the salting-out method13. The homozygous deletion of GSTM1 and GSTT1 genes was verified by Polymerase Chain Reaction (PCR), including the GSTP1 gene amplification as an internal control of the PCR reaction, and genotypes were considered to be present only when the band of the corresponding fragment was identified along with the control amplification band.
Multiplex PCR was carried out in a mixture of 2.5 mL of DMSO, 5 mL of Buffer 10x Taq buffer with (NH4)2SO4, 2.5 mM of MgCl2, 2.5 mM of dNTPs, 0.8 pMol of each primer, 100 ng of genomic DNA and 3U of Taq DNA polymerase (Fermentas®) for a 50 µL reaction. The reaction involved initial denaturation of 5 minutes at 95°C, 6 cycles of Touchdown (decreasing 1°C/cycle at annealing temperature) at 94°C for 1 minute, 59 - 54°C for 2 minutes, 72°C for 1 minute, followed by 30 cycles of 94°C for 1 minute, 55°C for 1 minute, 72°C for 1 minute with a final extension at 72°C for 5 minutes.
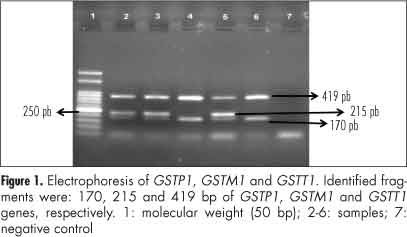
The presence or absence of GSTT1 and GSTM1 was detected by the presence or absence of a fragment of 419 and 215 bp, respectively. The size of the generated fragment corresponding to the internal control of the reaction (GSTP1) was 176 bp and was present in all amplifications. The following primers were used: 5'-GGT GGA AGG GAC AAG GTA GT-3' and 5'-TAA AGT GCT GAC CTG GGA AG-3' for GSTT1, primers 5'-GAA CTC CCT GAA AAG CTA AGC-3' and 5'-GTT GGG GCT CAA ATA TAC GGT GG-3' for GSTM1 and primers 5'-ACC CCA GGG CTC TAT GGG AA-3' and 5'-TAG GGG CAC AAG AAG CCC CT-3' for GSTP1. After electrophoresis on 2% agarose gel containing 5 mL of Ethidium Bromide, the fragments were visualized on an ultraviolet transilluminator and compared with molecular weight marker (50 bp, Ludwig, Biotec Ltd., Porto Alegre, RS) (Figure 1).
Data analysis
Information from questionnaires and laboratory tests were categorized, coded, entered and analyzed in a database constructed through the Statistical Package for Social Sciences (SPSS) software, release 20.0 (SPSS Inc., Chicago, USA). Descriptive statistics and univariate comparisons were performed. In the bivariate analyses, χ2 or Fisher's exact test were used to verify the presence of associations between data regarding differences between proportions, and Student's t-test was used for differences between means. Differences were considered significant if the p-value did not exceed 0.05. Odds ratios (OR) and 95% confidence intervals (95%CI) were also calculated.
Results
Epidemiological analysis
A total of 98 women (49 cases and 49 controls) were enrolled in this study. The mean age at the time of sample collection of patients included as cases was 54.9 years (±10.9), ranging from 36 to 82 years and the mean age of patients enrolled as controls was 53.8 years (±10.1), ranging from 35 to 83 years. Regarding ethnicity, 77.6 and 85.7% were white, respectively, in cases and controls. Among the women who had no children, the frequency of breast cancer was 87.5%. Other factors such as smoking, use of hormone replacement therapy and contraceptive use were not associated with risk of breast cancer (Table 1).
The mean age at the diagnosis of breast cancer was 52.5 years (±10.9) ranging from 33 to 80 years old. More than 55% of the patients had their diagnosis after 50 years old. Only 18.4% of the patients were diagnosed in Stage I, whereas 20.4% of the patients were diagnosed in Stage IV. Of all the 49 patients with breast cancer in the study, only 3 (6.1%) did not undergo chemotherapy. The most commonly used regimens were ACD (Adriamycin®, cyclophosphamide, docetaxel) and FAC (Fluorouracil, Adriamycin®, cyclophosphamide) in 32.6 and 30.4% of the women, respectively, followed by ACT (Adriamycin®, cyclophosphamide, Taxol®) and AC (Adriamycin®, cyclophosphamide) in 17.4 and 10.9%, respectively. The remaining 8.7% were treated with Xeloda® and CMF (cyclophosphamide, methotrexate and fluorouracil). Regarding the tumors, approximately 71.5% of the patients had estrogen-positive receptors, and, consequently, it was possible to use drugs that were potentially blocking for these receptors, such as Tamoxifen, which was used for 88.6% of the patients with this status.
Analysis of genes GSTT1 and GSTM1
The frequency of homozygous deletion of GSTM1 and GSTT1 genes among the 98 patients was 17.3 and 47.9%, respectively. The deletion of the isolated or combined genes in the different groups, cases and controls, can be seen in Table 2. No association was found between the pattern of gene deletions with age at diagnosis, nulliparity, smoking, oral contraceptive use, tumoral stage, familiar history and breastfeeding duration.
We observed that the homozygous deletion of GSTM1 gene was significantly associated with the development of breast cancer when compared with controls (Table 2). A total of 7 (7.1%) women had double deletions; of these, 5 (71.4%) had breast cancer. Among the 17 (34.7%) patients with a family history of breast cancer, 4 (23.5%) and 7 (41.2%) had a deletion of the GSTM1 and GSTT1 genes, respectively.
Discussion
Worldwide, epidemiological and molecular studies have been performed in order to better understand the development of breast cancer. Risk factors related to hormones, such as age at menarche, menopause and first pregnancy may influence the development of this disease due to the time of exposure to endogenous estrogen. Moreover, factors such as use of oral contraceptives and hormone replacement therapy have shown an increased exposure to exogenous estrogen10. However, in the study population, these characteristics were not classified as being a risk for the development of breast cancer, with similar results having been reported by Saxena et al.14.
Several studies have reported that the risk of breast cancer increases with age due to the longer exposure to endogenous hormone; therefore, it is a relatively rare disease before 35 years of age2. Regarding ethnicity, in this study, 77.6% of the women with breast cancer were Caucasians; these results corroborate others described in the literature, indicating that Caucasian women are at higher risk of developing breast cancer14-16.
Nulliparity and late primiparity have been described by several authors as risk factors for breast cancer development, for the onset of the first pregnancy helps the breast cell maturation process, making them potentially more protected against the action of carcinogens. Furthermore, there are indications of the important benefits of breastfeeding for women's health, resulting in a lower risk of breast cancer17. In this study, 87.5% of the nulliparous women had breast cancer. We are not be able to identify if the nulliparity was due to the illness or due to the treatment. A case-control study carried out with an Indian population that investigated 413 cases of breast cancer and 410 controls showed that the risk of breast cancer was lower among women who had had at least one pregnancy14.
Moraes et al.18 studied a population of women with breast cancer in the central region of Rio Grande do Sul and found that approximately 69.6% of the patients had tumors with positive estrogen receptors. Another study showed that 60.9% of the tumors had estrogen receptors19. The present study showed similar results, in which 71.5% of the patients had estrogen receptors in their tumors. It is extremely important to verify the presence of receptors to establish a correct treatment approach, as the tumor with estrogen receptors develops more rapidly in the presence of the hormone, which acts as a potent promoter of mitosis in cancer cells. Thus, it is necessary the use of drugs to competitively block these receptors, such as Tamoxifen, thereby preventing tumor growth and recurrence, as the treatment is long and can last up to five years19.
The homozygous deletions of GSTM1 and GSTT1 genes are common and result in a complete loss of enzymatic activity, so that individuals who carry them are more susceptible to developing diseases related to exposure to carcinogens10,20. The present study disclosed absence of GSTM1 and GSTT1 genes in 17.3 and 47.9% of the study participants, respectively. According to Reis, GSTM1 and GSTT1 genes have polymorphisms of which 10 and 50% respectively of the individuals in several populations have homozygous deletion21. Anton et al.22 verified the presence of GSTM1 and GSTT1 gene polymorphisms in a population similar to that evaluated in this study and found that 44.4% of the women had homozygous deletion of GSTM1 and 46.3%, of GSTT1. The difference in the frequency of the GSTT1 deletion may be related to the number of investigated subjects, and especially with the methodology used in this study, which differed by the absence of an internal control in the PCR.
In this study, the women with a higher proportion of deletion of the GSTM1 and GSTT1 genes, 58.8 and 61.7%, respectively, had breast cancer. Women with deletion of these genes have a three-fold higher susceptibility to breast cancer, especially in the premenopausal period11. Some authors found no significant association between the isolated deletion of GSTM1 and GSTT1 genes and the risk of breast cancer21,22. Morais et al.7 found approximately 35% of cases with GSTM1 gene deletion and only 14% of the GSTT1 gene, but could not associate the deletion with the risk of breast cancer.
Hayes and Pulford23 suggest that individuals with homozygous deletion of GSTs, mainly the GSTM1 gene, have a high risk of developing several types of cancer. In this study, the homozygous deletion of GSTM1 gene in the case group, irrespective of familial and sporadic cases, was associated with susceptibility to breast cancer. The absence of GSTM1 gene was also associated with the risk of developing breast cancer in other studies12,24. On the other hand, Khedhaier et al.25 found no significant difference between cases and controls in a Tunisian population, when correlated to the absence of this gene. These differences in results between the studies may be explained by variations in sample size, differences in study populations, inclusion criteria for controls, environmental factors, dietary habits and the methodology used for genetic analysis.
Chen et al. carried out a meta-analysis of 17,254 cases and 21,163 controls and observed that the risk of breast cancer was significantly higher when associated with the absence of GSTT1 gene24. However, few studies have associated GSTT1 gene deletion with an increased risk of cancer susceptibility14,22.
Anton et al.22, when analyzing a population from Santa Cruz do Sul, found that of the total number of women who had double homozygous deletion of genes GSTM1/GSTT1, 70% were breast cancer patients, showing that the double homozygous deletion is associated with susceptibility to breast cancer. In this study, the frequency was similar (71.4%), although there was no statistical association between the double deletion with susceptibility to breast cancer. This difference in results is probably related to the difference obtained in the frequency of GSTT1 gene deletion, which was higher in the study by Anton et al.22, when compared to the frequency observed in the present study. Khedhaier et al.25 could not associate the double deletion with breast cancer, but observed that the deletion was higher in cases than in controls, which is in agreement with what was observed in the present study.
If the exogenous factors that increase the risk of breast cancer can be avoided in individuals with susceptible genotypes, then, potentially, a significant proportion of breast cancer cases can be prevented14.
The results of this study suggest that the homozygous deletion of GSTM1 gene is associated with an increased susceptibility to breast cancer in this population. However, further studies are important, including one with a larger population, so these findings can be confirmed.
Acknowledgements
We thank our colleagues and professors of the Laboratory of Genetics and Biotechnology of Universidade Santa Cruz do Sul and the technical staff of the Center for Integrative Oncology of Hospital Ana Nery for patient recruitment and help in sample collection.
Received: 10/08/2013
Accepted with modifications: 12/13/2013
Conflict of interests: none.
Laboratório de Biotecnologia, Universidade de Santa Cruz do Sul - UNISC - Santa Cruz do Sul (RS), Brazil
- 1. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893-917.
-
2Brasil. Ministério da Saúde. Instituto Nacional de Câncer [Internet]. Estimativa 2012: incidência de câncer no Brasil. Rio de Janeiro: INCA: 2012 [cited 2012 Nov 8]. Available from: http://www.inca.gov.br/estimativa/2012/estimativa20122111.pdf
- 3. Tiezzi DG. [Epidemiology of breast cancer]. Rev Bras Ginecol Obstet. 2009;31(5):213-5.
- 4. González DM, Cabrera SJ, Hurlé AD. Farmacogenética en oncología. Med Clin. 2008;131(5):184-95.
- 5. Townsend DM, Tew KD. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene. 2003;22(47):7369-75.
- 6. Honma HN, De Capitani EM, Perroud MW Jr, Barbeiro AS, Toro IF, Costa DB, et al. Influence of p53 codon 72 exon 4, GSTM1, GSTT1 and GSTP1*B polymorphisms in lung cancer risk in a Brazilian population. Lung Cancer. 2008;61(2): 152-62.
- 7. Morais LM, Cardoso Filho C, Lourenço GJ, Shinzato JY, Zeferino LC, Lima CS, et al. Características mamográficas do câncer de mama associadas aos polimorfismos GSTM1 E GSTT1. Rev Assoc Med Bras. 2008;54(1):61-6.
- 8. Romero A, Martín M, Oliva B, De la Torre J, Furio V, De la Hoya M, et al. Glutathione S-transferase P1 c.313A > G polymorphism could be useful in the prediction of doxorubicin response in breast cancer patients. Ann Oncol. 2012;23(7):1750-6.
- 9. Cho SG, Lee YH, Park HS, Ryoo K, Kang KW, Park J, et al. Glutathione S-transferase mu modulates the stress-activated signals by suppressing apoptosis signal-regulating kinase 1. J Biol Chem. 2001;276(16):12749-55.
- 10. Torresan C, Oliveira MM, Torrezan GT, de Oliveira SF, Abuázar CS, Losi-Guembarovski R, et al. Genetic polymorphisms in oestrogen metabolic pathway and breast cancer: a positive association with combined CYP/GST genotypes. Clin Exp Med. 2008;8(2):65-71.
- 11. Park SK, Kang D, Noh DY, Lee KM, Kim SU, Choi JY, et al. Reproductive factors, glutathione S-transferase M1 and T1 genetic polymorphism and breast cancer risk. Breast Cancer Res Treat. 2003;78(1):89-96.
- 12. Strange RC, Jones PW, Fryer AA. Glutathione S-transferase: genetics and role in toxicology. Toxicol Lett. 2000;112-113: 357-63.
- 13. Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215.
- 14. Saxena A, Dhillon VS, Raish M, Asim M, Rehman S, Shukla NK, et al. Detection and relevance of germline genetic polymorphisms in glutathione S-transferases (GSTs) in breast cancer patients from northern Indian population. Breast Cancer Res Treat. 2009;115(3):537-43.
- 15. Reding KW, Chen C, Lowe K, Doody DR, Carlson CS, Chen CT, et al. Estrogen related genes and their contribution to racial differences in breast cancer risk. Cancer Causes Control. 2012;23(5):671-81.
- 16. Sakoda LC, Blackston CR, Xue K, Doherty JA, Ray RM, Lin MG, et al. Glutathione S-transferase M1 and P1 polymorphisms and risk of breast cancer and fibrocystic breast conditions in Chinese women. Breast Cancer Res Treat. 2008;109(1):143-55.
- 17. Rea MF. Os benefícios da amamentação para a saúde da mulher. J Pediatr (Rio J). 2004;80(5 Supl):S142-6.
- 18. Moraes AB, Zanini RR, Turchiello MS, Riboldi J, Medeiros LR. Estudo da sobrevida de pacientes com câncer de mama atendidas no hospital da Universidade Federal de Santa Maria, Rio Grande do Sul, Brasil. Cad Saúde Pública. 2006;22(10):2219-28.
- 19. Lizard-Nacol S, Coudert B, Colosetti P, Riedinger JM, Fargeot P, Brunet-Lecomte P. Glutathione S-transferase M1 null genotype: lack of association with tumour characteristics and survival in advanced breast cancer. Breast Cancer Res. 1999;1(1):81-7.
- 20. Song DK, Xing DL, Zhang LR, Li ZX, Liu J, Qiao BP. Association of NAT2, GSTM1, GSTT1, CYP2A6, and CYP2A13 gene polymorphisms with susceptibility and clinicopathologic characteristics of bladder cancer in Central China. Cancer Detect Prev. 2009;32(5-6):416-23.
- 21. Reis M. Farmacogenética aplicada ao câncer: quimioterapia individualizada e especificidade molecular. Medicina (Ribeirão Preto). 2006;39(4):577-86.
- 22. Anton EM, Renner JD, Valim AR, Dotto ML, Possuelo LG. Avaliação epidemiológica da influência dos genes GSTM1 e GSTT1 na susceptibilidade ao câncer de mama em mulheres atendidas em um hospital do sul do Brasil: um estudo piloto. Rev AMRIGS. 2010;54(4):411-5.
- 23. Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30(6):445-600.
- 24. Chen XX, Zhao RP, Qiu LX, Yuan H, Mao C, Hu XC, et al. Glutathione S-transferase T1 polymorphism is associated with breast cancer susceptibility. Cytokine. 2011;56(2):477-80.
- 25. Khedhaier A, Remadi S, Corbex M, Ahmed SB, Bouaouina N, Mestiri S, et al. Glutathione S-transferases (GSTT1 and GSTM1) gene deletions in Tunisians: susceptibility and prognostic implications in breast carcinoma. Br J Cancer. 2003;89(8):1502-7.
Publication Dates
-
Publication in this collection
03 Feb 2014 -
Date of issue
Dec 2013
History
-
Received
10 Aug 2013 -
Accepted
13 Dec 2013