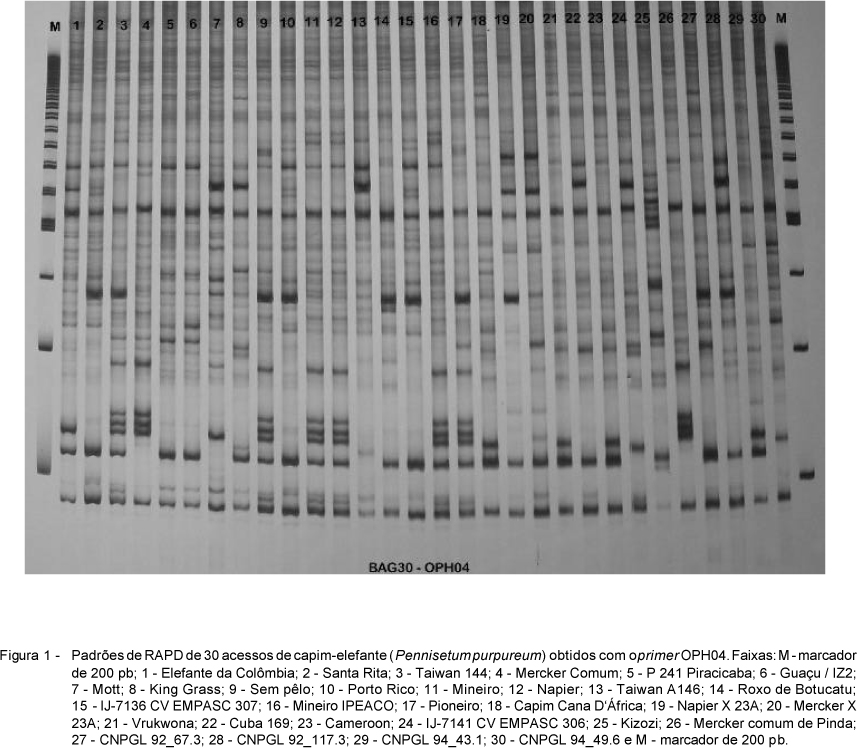
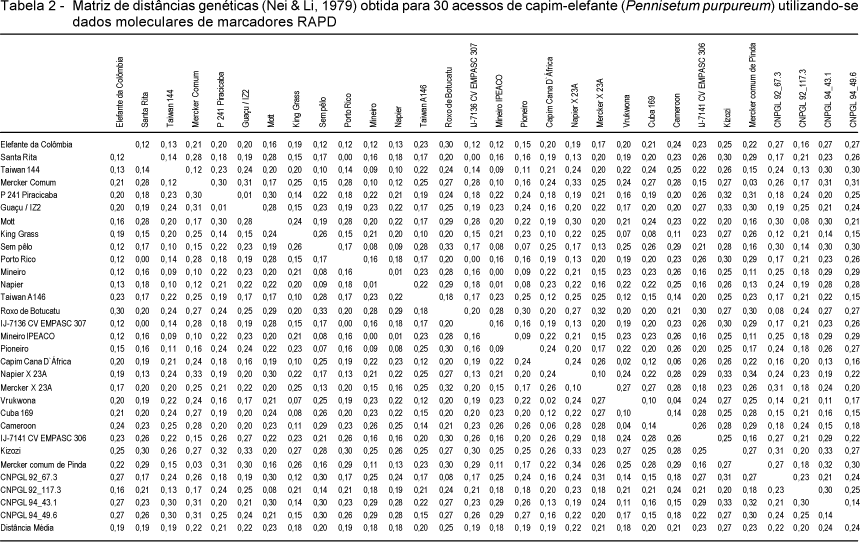
This study aimed to estimate the genetic diversity among Elephantgrass accessions using molecular markers. DNA samples of 30 accessions were obtained to perform PCR amplification using RAPD technique. Twenty random primers were used to obtain 88 high intensity and reproducible DNA bands (64 were polymorphic and 24 were monomorphic). Accessions were grouped by genetic distance estimates using UPGMA clustering. Genetic distances among Porto Rico, Santa Rita, IJ-7136 cv EMPASC 307; Mineiro and Mineiro Ipeaco accessions were null, suggesting that they are genetically the same although labeled with different names. Genetic distances were minimum between the pairs of accessions Napier and Mineiro or Mineiro Ipeaco (0,01); P 241 Piracicaba and Guaçu / IZ2 (0,01); Capim Cana D'África and Vrukwona (0,02); Merker Comum and Merker Comum de Pinda (0,03) and indicate that their genotypes are closely related. The largest genetic distance (0,34) was observed between the Mercker Comum de Pinda and Napier X 23A accessions. Overall, the results indicate genetic variability among accessions and suggest that genetic distance may be used as an auxiliary criterion of selection in breeding programs of elephantgrass genotypes.
genetic variability; molecular markers; Pennisetum purpureum; RAPD