Abstract
English grain aphid (EGA, Sitobion avenae Fabricius) is an important pest in wheat (Triticum aestivum L.). To develop EGA-resistant varieties, introducing the desirable genes from related species is regarded as an efficient avenue. In this study, the F1, F2 and F3 plants derived from the cross of EGA-susceptible wheat-Psathyrostachys huashanica Keng ex Kuo amphiploid (PHW-SA, AABBDDNsNs) and EGA-resistant triticale (Zhongsi 828, AABBRR) were analyzed for EGA resistance. Consequently, PHW-SA was moderately susceptible while Zhongsi 828 and their F1 hybrids were immune, suggesting that the resistance is dominant. All the F2 plants showed high resistance or immunity over two years, indicating that EGA resistance genes are more likely carried by the rye (Secale cereale L.) chromosomes rather than the genomes A or B of Zhongsi 828. In the F3 generation, 25 of 239 lines became susceptible. Giemsa C-banding patterns revealed that these F3 lines had 38-40 chromosomes, including complete rye genome except 5R (and 2R in five lines). Genomic in situ hybridization analysis confirmed this result. During meiosis, all the chromosomes formed bivalents. Six bivalents in 20 lines and five bivalents in five lines were characterized from rye. In contrast, their F2 parental lines had 42 chromosomes (21 bivalents), containing 1R-7R of rye. No P. huashanica chromosomes were detected. Therefore, we propose that the rye chromosome 5R may be related to EGA resistance.
chromosome constitutions; genetic resources; wide cross
GENETICS AND PLANT BREEDING
Cytogenetic identification of wheatPsathyrostachys huashanica amphiploid × triticale progenies for English grain aphid resistance
Quan XieI,II; Houyang KangI; Debbie Louise SparkesIII; Shan TaoI; Zhiqin HuI; Lili XuI; Xing FanI; Lina ShaI; Haiqin ZhangI; Yi WangI; Jian ZengIV; Yonghong ZhouI,II,* * Corresponding author < zhouyhdavid@126.com>
ISichuan Agricultural University/Triticeae Research Institute, Wenjiang, Chengdu 611 130 Sichuan China
IISichuan Agricultural University/Key Laboratory of Crop Genetic Resources and Improvement Ministry of Education
IIIUniversity of Nottingham/Division of Plant and Crop Sciences, Sutton Bonington Campus, Leicestershire LE12 5RD Loughborough UK
IVSichuan Agricultural University/College of Resources and Environment
ABSTRACT
English grain aphid (EGA, Sitobion avenae Fabricius) is an important pest in wheat (Triticum aestivum L.). To develop EGA-resistant varieties, introducing the desirable genes from related species is regarded as an efficient avenue. In this study, the F1, F2 and F3 plants derived from the cross of EGA-susceptible wheatPsathyrostachys huashanica Keng ex Kuo amphiploid (PHW-SA, AABBDDNsNs) and EGA-resistant triticale (Zhongsi 828, AABBRR) were analyzed for EGA resistance. Consequently, PHW-SA was moderately susceptible while Zhongsi 828 and their F1 hybrids were immune, suggesting that the resistance is dominant. All the F2 plants showed high resistance or immunity over two years, indicating that EGA resistance genes are more likely carried by the rye (Secale cereale L.) chromosomes rather than the genomes A or B of Zhongsi 828. In the F3 generation, 25 of 239 lines became susceptible. Giemsa C-banding patterns revealed that these F3 lines had 3840 chromosomes, including complete rye genome except 5R (and 2R in five lines). Genomic in situ hybridization analysis confirmed this result. During meiosis, all the chromosomes formed bivalents. Six bivalents in 20 lines and five bivalents in five lines were characterized from rye. In contrast, their F2 parental lines had 42 chromosomes (21 bivalents), containing 1R7R of rye. No P. huashanica chromosomes were detected. Therefore, we propose that the rye chromosome 5R may be related to EGA resistance.
Keywords: chromosome constitutions, genetic resources, wide cross
Introduction
Aphid (Hemiptera: Aphididae) is one of the major pests of cereal crops worldwide, especially in temperate regions. They cause a significant loss of yield by consuming the photoassimilates in plant sap and by functioning as the vectors transmitting over 250 plant viruses such as barley yellow dwarf virus (BYDV) (Nault, 1997). In wheat (Triticum aestivum L.), the English grain aphid (EGA) is the most important pest, known as an ear feeder in summer. It distributes extensively in Northern and Western China almost every year, and can give rise to a remarkable reduction in wheat yield (up to 70 %) as well as flour quality under heavy infestation (Shi et al., 2009).
As a critical part of the integrated pest management (IPM), the development of aphid-resistant wheat varieties will be a sustainable way in a long term. Many aphid resistance genes have been found and located in wheat. For example, genes Dn1, Dn2, Dn5, Dn6 and DnX located on chromosome 7DS, Dn4 on 1DS, Dn8 on 7DL and Dn9 on 1DL express resistance to the Russian wheat aphid (Liu et al., 2005; Tyrka and Chelkowski, 2004). Genes Gb3 and Gbz on chromosome 7DL function against the greenbug (Weng and Lazar, 2002; Zhu et al., 2004), whilst a single dominant gene RA-1 on chromosome 6AL is identified against EGA (Liu et al., 2012).
Attention has been paid to broadening genetic variation of crop plants over the past decades. For wheat, there are many related species present in Triticeae. They have the potential to contribute to the development of wheat cultivars with aphid resistance. The resistance to aphid races has been found in some species of Agropyron (Tremblay et al., 1989), Avena (Weibull, 1986), Elymus (Tremblay et al., 1989), Hordeum (Weibull, 1987), Secale (Anderson et al., 2003), Triticum (Lage et al., 2004) and several accessions of triticale (Webster, 1990).
In the present study, a cross was conducted between the EGA-susceptible female parent, wheatPsathyrostachys huashanica Keng ex Kuo amphiploid 'PHW-SA' (AABBDDNsNs) and the EGA-resistant male parent, hexaploid triticale 'Zhongsi 828' (AABBRR). The offspring family (F1 to F3 generations) was analyzed for EGA resistance. Interestingly, 25 F3 lines became high susceptible as compared to their F2 parental lines and sibs carrying resistance to EGA. The genomic constitutions of these lines were then characterized cytogenetically.
Materials and Methods
Plant materials
A cross between the EGA-susceptible wheatP. huashanica amphiploid 'PHW-SA' (2n = 8x = 56, AABBDDNsNs) and the EGA-resistant triticale cultivar 'Zhongsi 828' (2n = 6x = 42, AABBRR) was obtained in 2008 (Kang et al., 2011). The F1 plants were selfed for three generations. As a result, 26 F2 lines and 239 F3 lines were produced in 2010 and 2011, respectively. The individuals of F1 and F2 generations were cultivated in 2011 as well. A rye cultivar 'Qinling' (2n = 14, RR) and P. huashanica (2n = 14, NsNs) were used as the probes for genomic in situ hybridization. Common wheat 'Chinese Spring' ('CS', 2n = 42, AABBDD) was used as the blocker. These plant materials went through the cropping seasons under natural field conditions without any fertilizer or pesticide.
EGA resistance evaluation
The resistance of F1, F2 and F3 plants to EGA was evaluated in two cropping seasons. Only F2 plants were investigated in 2010, and all the F1, F2 and F3 plants observed in 2011. Evaluation of EGA resistance was carried out under the natural infection conditions as this aphid appears regularly every year. For each line, three spikes of the main shoots were cut at the milky stage and the number of the aphids was counted. The ratios of the average aphid number per spike of each line and the average aphid number per spike of all the lines were calculated, termed as aphid indexes. A 0 (immune) 6 (highly susceptible) scale was employed to denote the infection severity according to the Painter's method (1951). That is, 0 = immunity, aphid index being 0; 1 = high resistance, aphid indexes ranging from 0.01 to 0.30; 2 = moderate resistance, aphid indexes ranging from 0.31 to 0.60; 3 = low resistance, aphid indexes ranging from 0.61 to 0.90; 4 = low susceptibility, aphid indexes ranging from 0.91 to 1.20; 5 = moderate susceptibility, aphid indexes ranging from 1.21 to 1.50; 6 = high susceptibility, aphid indexes being more than 1.50.
Giemsa C-banding
The Giemsa C-banding technique was adopted to identify individual alien chromosomes. The seeds were germinated at 25 ºC, and their roots that were 1 to 2 cm in length were collected. The roots were then fixed in the fresh Carnoy's fixative I (3 volumes of 100 % ethanol + 1 volume of glacial acetic acid) solution for 24 h, after pretreatment in ice-cold water for about 20 h, followed by hydrolyzation in 0.2 mol L1 HCl for 34 h. The meristem cells were squashed in 45 % acetic acid and the cover glasses were removed through freezing in liquid nitrogen. Giemsa C-banding was performed using the methods described by Gill et al. (1991) as revised by Wang et al. (2011). The characterization of S. cereale and P. huashanica chromosomes was made by matching the standard patterns demonstrated by Gill and Kimber (1974), and Zhang et al. (2009), respectively.
Meiotic analysis
Meiotic analysis of pollen mother cells (PMCs) was conducted to understand the chromosome pairing. The growing spikes were removed when the flag leaf sheathes reached around 5 cm length, followed by immediate fixation in Carnoy's fixative II (six volumes of 100 % ethanol + three volumes of chloroform + one volume of glacial acetic acid) solution for 24 h. One of three anthers in a floret during meiosis was squashed in a drop of modified carbol fuchsin. Chromosome pairing was analyzed based on 50 PMCs.
Genomic in situ hybridization (GISH)
The genomic constitutions of somatic cells and PMCs were further identified by GISH. The procedures of chromosome preparation were described earlier. For meiotic analysis, the remaining two anthers were used here. The slides were air-dried, post-fixed in 4 % paraformaldehyde for 10 min, denatured in 70 % deionized formamide at 80 ºC for 2 min, and then dehydrated in ethanol series (-20 ºC, 5 min each in 70 %, 95 % and 100 %).
The genomic DNA of plant materials was isolated by the hexadecyltrimethylammonium bromide (CTAB) method (Doyle and Doyle, 1990). The total genomic DNA of rye (RR) and P. huashanica (NsNs) were labeled, as the probes with digoxigenin-11-dUTP, by nick translation following the manufacturer protocol (Roche, Mannheim, Germany). The total genomic DNA of CS was autoclaved at 115 ºC for 10 min as the blocker (200500 bp). A sample of 30 µL hybridization solution, which included 50 % formamide, sodium chloride-sodium citrate (SSC) 2X buffer, 10 % dextran sulfate, 0.17 mg mL1 herring sperm DNA (200500 bp), 50 ng probe DNA and 3,000 ng CS DNA, was used for each slide. GISH was performed by the method described by Chen et al. (1996) and modified by Wang et al. (2011). After being counterstained by propidium iodide (PI, 1.5 µg mL1) (Vector, California, USA), the hybridization signals on chromosomes were visualized with a fluorescence microscope equipped with green and blue filters. Images were recorded with a CCD (charge-coupled device) camera.
Results
EGA resistance evaluation
The progenies of the F2 generation in 2010 and F1, F2 and F3 generations in 2011, together with the female parent PHW-SA and male parent Zhongsi 828, were screened for the reaction types to EGA (Table 1). PHW-SA was moderately susceptible whereas Zhongsi 828, F1 and F2 plants were immune or highly resistant. Out of 239 F3 lines, 25 from five F2 parental lines were highly susceptible to EGA (Figure 1). The other F3 lines, however, exhibited high resistance or immunity.
Mitotic analysis
The mitotic metaphase cells in root tips of the F3 lines with high susceptibility to EGA, their F2 parental plants and sibs were analyzed for the chromosome number and compositions in order to determine the genetic difference among them (Table 2). The chromosome number of F2 parental lines was consistently 42. In 25 F3 lines, 20 had 40 chromosomes and the rest contained only 38 chromosomes.
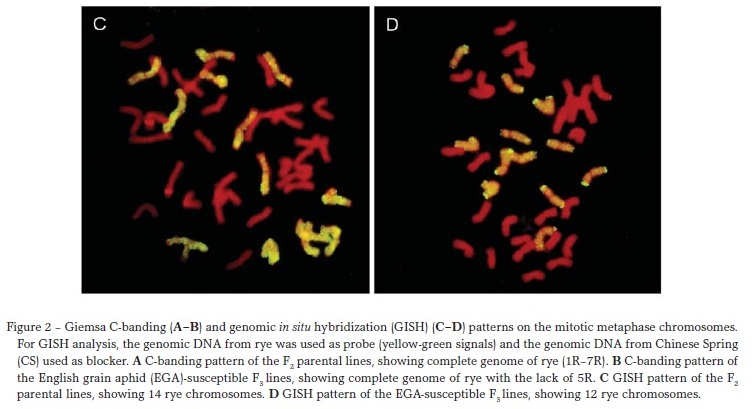
Giemsa C-banding and GISH were used to characterize the chromosome compositions of these lines. C-banding patterns indicated that five F2 parental lines included complete R genome of rye (Figure 2A). Among 25 F3 lines, 20 had each pair of rye genome except 5R (Figure 2B) and five lacked both 5R and 2R. Eight F3 sibs from the same F2 parents were selected randomly, and most of them had a complete rye genome (1R7R) and the remaining ones had 1R or 2R lost. There were no P. huashanica chromosomes found in all the examined combinations.
GISH confirmed the results of Giemsa C-banding (Figure 2CD). Fourteen chromosomes of rye were denoted by yellow-green fluorescence in five F2 parental lines (Figure 2C). Twelve rye chromosomes were present in 20 F3 lines (Figure 2D), and ten were detected in five lines. No hybridization signals were found when the DNA of P. huashanica was used as the probe.
Meiotic analysis
PMCs in these lines were further observed to understand their chromosome behavior during meiosis (Figure 3). Five F2 parental lines formed 21 bivalents in the most of PMCs at meiotic metaphase I (MI) (Figure 3A). Twenty and 19 bivalents were observed in 20 (Figure 3B) and five target F3 lines. At anaphase I, one of F2 parental lines yielded a pair of lagging chromosomes, and a number of micronuclei were produced at telophase II. The other F2 parents and F3 plants progressed normally.
The rye chromosomes during meiosis were visualized by GISH. Seven bivalents were consistently seen in the F2 parents at MI, clustering with the wheat chromosomes at the equatorial plate (Figure 3C). Six and five bivalents from rye were identified in 20 and five target F3 lines, suggesting that these chromosomes were homologous (Figure 3D). As the cells reached the anaphase I, these bivalents were able to separate regularly. Overall, it can be summarized that 20 of 25 F3 lines showing high susceptibility to EGA were nullisomic for 5R and the remaining lines lacked both 2R and 5R, compared with their F2 parental lines that contained the complete rye genome.
Discussion
The F1 hybrids of PHW-SA (2n = 8x = 56, AABBDDNsNs) and Zhongsi 828 (2n = 6x = 42, AABBRR) have 49 chromosomes, comprising AABBDNsR (Kang et al., 2011). After selfing, three genomes (D, Ns and R) might undergo elimination since they could not pair and separate normally in meiosis without their homologous chromosomes. In fact, there were 42 chromosomes in five F2 parental lines. No chromosomes of P. huashanica were found but the complete rye genome (1R7R) was observed in these lines. Additionally these chromosomes formed 21 bivalents at MI. This suggests that D and Ns genomes had been lost while R genome had been doubled during propagating. Fourteen out of 21 bivalents, theoretically, belong to the A and B genomes of wheat and the remaining ones are the chromosomes of rye. In other words, the F2 parental lines may be considered as new triticale (2n = 6x = 42, AABBRR). The absence of wheat D genome could be beneficial because this can reduce the confounding effects of genes on the genome. For example, several major genes conferring aphid resistance have been located on the D genome (Liu et al., 2005; Weng and Lazar, 2002; Zhu et al., 2004). The other aphid resistance genes could thus be detected easily without the effect of these major ones.
Screening for resistance to EGA revealed that the female parent PHW-SA was susceptible whereas the male parent Zhongsi 828 was immune. Their F1 hybrids showed immunity. One can deduce the existence of certain dominant resistance gene(s) in Zhongsi 828 and successful transmission of those to the hybrids. In the F2 segregating population all the plants remained at high resistance or immunity to the aphids over two years. This indicates that these genes are less likely on the genome A or B of Zhongsi 828 as there should have been susceptible genotypes in F2 plants after recombination. Further, only 25 lines in the F3 generation became highly susceptible to EGA. In contrast to their F2 parental lines and sibs, they consistently lacked chromosome 5R of rye. It may therefore be speculated that chromosome 5R of rye functions against EGA.
To date about seven aphid resistance genes have been identified in rye. Two greenbug resistance genes, Gb2 and Gb6, and one Russian wheat aphid resistance gene Dn7 are located on 1RS, and four greenbug resistance genes Dnx (Dnr1, Dnr2, Dnr3, and Dnr4) located on 1RL, 3RS, 4R and 7R, respectively (Fritz et al., 1999; Lapitan et al., 2007; Marais et al., 1994; Nkongolo et al., 1996; Tyrka and Chelkowski, 2004). Numerous translocations, additions and synthetic triticale have been developed for transfer of these genes to wheat (Hesler, 2005; Marais et al., 1994; Nkongolo et al., 2009, 2011), and a number of molecular maps are available (Anderson et al., 2003; Fritz et al., 1999; Lapitan et al., 2007; Tyrka and Chelkowski, 2004). However, few studies have focused on EGA in spite of its tremendous damage in wheat worldwide. Only one dominant gene responsible for EGA resistance was reported on chromosome 6AL of wheat (Liu et al., 2012). In the present study, it is elucidated that chromosome 5R of rye may carry major gene(s) for EGA resistance. These gene(s) should be different from those reported on 1R, 3R, 4R and 7R. The mechanism of these genes from rye against aphids remains unknown but it is possibly associated with high concentrations of hydroxamic acids (mainly DIBOA) in rye plants (Niemeyer et al., 1992).
Rye has been used widely as a resource for broadening the genetic base of wheat. Over 39 resistance genes against diseases and pests have been found in rye (Tyrka and Chelkowski, 2004). Here we report the existence of the putative genes conferring EGA resistance on chromosome 5R of rye. This offers an opportunity to transfer them to wheat.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (31101151), the National Transgenic Major Project (2011zx08009001), the National High-tech R&D Program of China (863 Program) (2011AA100103-02), the Special Fund for Agro-Scientific Research in the Public Interest of China (201003021), the Natural Science Foundation for Young Scientists of Sichuan Province, China (2010JQ0046), and the Science and Technology Bureau and Education Bureau of Sichuan Province, China.
Received August 01, 2012
Accepted January 16, 2012
Edited by: Antonio Costa de Oliveira
- Anderson, G.R.; Papa, D.; Peng, J.; Tahir, M.; Lapitan, N.L.V. 2003. Genetic mapping of Dn7, a rye gene conferring resistance to the Russian wheat aphid in wheat. Theoretical and Applied Genetics 107: 1297-1303.
- Chen, Q.; Conner, R.L.; Laroche, A. 1996. Molecular characterization of Haynaldia villosa chromatin in wheat lines carrying resistance to wheat curl mite colonization. Theoretical and Applied Genetics 93: 679-684.
- Doyle, J.J.; Doyle, J.L. 1990. Isolation of plant DNA from fresh tissue. Focus 12: 13-15.
- Fritz, A.K.; Caldwell, S.; Worrall, W.D. 1999. Molecular mapping of Russian wheat aphid resistance from triticale accession PI 386156. Crop Science 39: 1707-1710.
- Gill, B.S.; Friebe, B.; Endo, T.R. 1991. Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (Triticum aestivum). Genome 34: 830-839.
- Gill, B.S.; Kimber, G. 1974. The Giemsa C-banded karyotype of rye. Proceedings of the National Academy of Sciences of the United States of America 71: 1247-1249.
- Hesler, L.S. 2005. Resistance to Rhopalosiphum padi (Homoptera: Aphididae) in three triticale accessions. Journal of Economic Entomology 98: 603-610.
- Kang, H.Y.; Zhong, M.Y.; Xie, Q.; Zhang, H.Q.; Fan, X.; Sha, L.N.; Xu, L.L.; Zhou, Y.H. 2011. Production and cytogenetics of trigeneric hybrid involving Triticum, Psathyrostachys and Secale Genetic Resources and Crop Evolution 59: 445-453.
- Lage, J.; Skovmand, B.; Andersen, S.B. 2004. Field evaluation of emmer wheat-derived synthetic hexaploid wheat for resistance to Russian wheat aphid (Homoptera: Aphididae). Journal of Economic Entomology 97: 1065-1070.
- Lapitan, N.L.V.; Peng, J.; Sharma, V. 2007. A high-density map and PCR markers for Russian wheat aphid resistance gene Dn7 on chromosome 1RS/1BL. Crop Science 47: 811-820.
- Liu, X.M.; Smith, C.M.; Friebe, B.R.; Gill, B.S. 2005. Molecular mapping and allelic relationships of Russian wheat aphid resistance genes. Crop Science 45: 2273-2280.
- Liu, X.L.; Yang, X.F.; Wang, C.Y.; Wang, Y.J.; Zhang, H.; Ji, W.Q. 2012. Molecular mapping of resistance gene to English grain aphid (Sitobion avenae F.) in Triticum durum wheat line C273. Theoretical and Applied Genetics 124: 287-293.
- Marais, G.F.; Horn, M.; du Torr, F. 1994. Intergeneric transfer (rye to wheat) of a gene(s) for Russian wheat aphid resistance. Plant Breeding 113: 265-271.
- Nault, L.R. 1997. Arthropod transmission of plant viruses: a new synthesis. Annals of the Entomological Society of America 90: 521-541.
- Niemeyer, H.M.; Copaja, S.V.; Barria, B.N. 1992. The Triticeae as sources of hydroxamic acids, secondary metabolites in wheat conferring resistance against aphids. Hereditas 116: 295-299.
- Nkongolo, K.K.; Haley, S.D.; Kim, N.S.; Michael, P.; Fedak, G.; Quick, J.S.; Peairs, F.B. 2009. Molecular cytogenetic and agronomic characterization of advanced generations of wheat x triticale hybrids resistant to Diuraphis noxia (Mordvilko): application of GISH and microsatellite markers. Genome 52: 353-360.
- Nkongolo, K.K.; Haley, S.D.; Quick, J.S.; Peairs, F.B. 2011. Registration of six wheatrye addition lines resistant to the Russian wheat aphid. Journal of Plant Registrations 5: 426-429.
- Nkongolo, K.K.; Lapitan, N.L.V.; Quick, J.S. 1996. Genetic and cytogenetic analyses of Russian wheat aphid resistance in triticale x wheat hybrids and progenies. Crop Science 36: 1114-1119.
- Painter, R.H. 1951. Insect Resistance in Crop Plants. Macmillan, New York, NY, USA.
- Shi, G.; Shang, X.; Wang, H.; Ma, X.; Hu, B.; Li, C. 2009. Responses of flour quality and dough rheological properties to Sitobion avenae F. inoculated in spring wheat. Acta Agronomica Sinica 35: 2273-2279 (in Chinese, with abstract in English).
- Tremblay, C.; Cloutier, C.; Comeau, A. 1989. Resistance to the bird cherry-oat aphid, Rhopalosiphum padi L. (Homoptera: Aphididae), in perennial gramineae and wheat x perennial gramineae hybrids. Environmental Entomology 18: 921-923.
- Tyrka, M.; Chelkowski, J. 2004. Enhancing the resistance of triticale by using genes from wheat and rye. Journal of Applied Genetics 45: 283-295.
- Wang, Y.; Yu, K.F.; Xie, Q.; Kang, H.Y.; Lin, L.J.; Fan, X.; Sha, L.N.; Zhang, H.Q.; Zhou, Y.H. 2011. The 3Ns chromosome of Psathyrostachys huashanica carries the gene(s) underlying wheat stripe rust resistance. Cytogenetic and Genome Research 134: 136-143.
- Webster, J.A. 1990. Resistance in triticale to the Russian wheat aphid (Homoptera: Aphididae). Journal of Economic Entomology 83: 1091-1095.
- Weibull, J. 1986. Screening for resistance against Rhopalosiphum padi (L.). I. Avena species and breeding lines. Euphytica 3: 993-999.
- Weibull, J. 1987. Screening for resistance against Rhopalosiphum padi (L.). II. Hordeum species and interspecific hybrids. Euphytica 36: 571-576.
- Weng, Y.; Lazar, D.M. 2002. Amplified fragment length polymorphism and simple sequence repeat based molecular tagging and mapping of greenbug resistance gene Gb3 in wheat. Plant Breeding 121: 218-223.
- Zhang, H.Q.; Yang, R.W.; Zhang, L.; Ding, C.B.; Zeng, J.; Zhou, Y.H. 2009. Genetic diversity and phylogeny in Hystrix (Poaceae, Triticeae) and related genera inferred from Giemsa C-banded karyotypes. Genetics and Molecular Biology 32: 521-527.
- Zhu, L.C.; Smith, C.M.; Fritz, A.; Boyko, E.V.; Flinn, M.B. 2004. Genetic analysis and molecular mapping of a wheat gene conferring tolerance to the greenbug (Schizaphis graminum Rondani). Theoretical and Applied Genetics 109: 289-293.
Publication Dates
-
Publication in this collection
14 May 2013 -
Date of issue
June 2013
History
-
Received
01 Aug 2012 -
Accepted
16 Jan 2012