Abstract
Transposable elements are dispersed repetitive DNA sequences that can move within the genome and are related to genome and chromosome evolution, adaptation, and speciation. The aim of this study was to characterize and determine the chromosomal location and accumulation of a Mariner-like element in populations of four phylogenetically related species of the Dichotomius (Luederwaldtinia) sericeus complex. Mapping of the isolated element was performed by fluorescent in situ hybridization in different populations of analyzed species. Characterization of the isolated element revealed a degenerated transposon, named DgmarMITE. This transposon is 496-bp-long, AT rich (57%), and contains 24 bp terminal inverted repeats. In situ mapping revealed presence of this element only in two out of four species analyzed. DgmarMITE sites were located in heterochromatic and euchromatic regions and varied in location and number on the karyotypes of Dichotomius (L.) gilletti and D. (L.) guaribensis across different populations. These results demonstrate differential accumulation of the DgmarMITE in genomes of these species, which is probably due to the occurrence of ectopic recombination and cross-mobilization of the element mediated by the transposase of closely related or unrelated transposable elements.
Keywords:
Mariner-like elements; cross-mobilization; chromosome evolution
Introduction
Mariner transposable elements are DNA transposons that exhibit broad diversity in their structure. Mariner elements are characterized by a size of about 1,300 bp, a single ORF (open reading frame) encoding a transposase, a conserved catalytic domain [DD(34)D] necessary for transposition, and two terminal inverted repeats (TIRs) of 28-30 bp flanked by a TA dinucleotide resulting from target site duplications (Robertson, 1995Robertson HM (1995) The Tcl-Mariner superfamily of transposons in animals. J Insect Physiol 41:99-105.; Robertson and Lampe, 1995Robertson HM and Lampe D (1995) Distribution of transposable elements in Arthropods. Annu Rev Entomol 40:333-357.; Plasterk et al., 1999Plasterk RHA, Izsvák Z and Ivics Z (1999) Resident aliens the Tc1/Mariner superfamily of transposable elements. Trends Genet 15:326-332.). During transposition, the encoded transposase recognizes the TIRs and catalyzes excision of the two DNA strands at the donor site and fusion of the element at another site in the genome (Lampe et al., 1996Lampe DJ, Churchill MEA and Robertson HM (1996) A purified Mariner transposase is sufficient to mediate transposition in vitro. EMBO J 15:5470-5479.).
The Mariner elements are probably the most widely distributed family of transposable elements (TEs) in nature, being represented in diverse taxa, such as rotifers, fungi, plants and vertebrates. Their wide distribution in metazoan species, including arthropods (Robertson and Lampe, 1995Robertson HM (1995) The Tcl-Mariner superfamily of transposons in animals. J Insect Physiol 41:99-105.; Wallau et al., 2014Wallau GL, Capy P, Loreto E and Hua-Van A (2014) Genomic landscape and evolutionary dynamics of Mariner transposable elements within the Drosophila genus. BMC Genomics 15:727.), is probably related to horizontal transfer events (Robertson 1995Robertson HM (1995) The Tcl-Mariner superfamily of transposons in animals. J Insect Physiol 41:99-105.; Robertson and Lampe, 1995Robertson HM (1995) The Tcl-Mariner superfamily of transposons in animals. J Insect Physiol 41:99-105.; Lampe et al., 2003Lampe DJ, Witherspoon DJ, Soto-Adames FN and Robertson HM (2003) Recent horizontal transfer of mellifera subfamily Mariner transposons into insect lineages representing four different orders shows that selection acts only during horizontal transfer. Mol Biol Evol 20:554-562.) which, for example, account for the presence of the Mariner_Tbel and Mariner1_BT families in phylogenetically distant species such as insects and mammals (Oliveira et al., 2012Oliveira SG, Bao W, Martins C and Jurka J (2012) Horizontal transfers of Mariner transposons between mammals and insects. Mobile DNA 3:1-6.). These elements have been found in a wide range of insects from different orders, including Diptera, Hemiptera, Hymenoptera, Lepidoptera, Orthoptera and Coleoptera (Robertson and Lampe, 1995Robertson HM (1995) The Tcl-Mariner superfamily of transposons in animals. J Insect Physiol 41:99-105.; Palacios-Gimenez et al., 2014Palacios-Gimenez OM, Bueno D and Cabral-de-Mello DC (2014) Chromosomal mapping of two Mariner-like elements in the grasshopper Abracris flavolineata (Orthoptera: Acrididae) reveals enrichment in euchromatin. Eur J Entomol 111:329-334.).
The existence of nonfunctional Mariner elements is common, including a large number of inactive copies in different genomes (Lohe et al., 1995Lohe AR, Moriyama EN, Lidholm DA and Hartl DL (1995) Horizontal transmission, vertical inactivation, and stochastic loss of Mariner-like transposable elements. Mol Biol Evol 12:62-72.). Some of those inactive elements, the miniature inverted repeat transposable elements (MITEs), do not encode the enzyme necessary for their transposition and therefore require the transposase of other elements for their mobilization (Kidwell, 2005Kidwell MG (2005) Transposable Elements. In: Gregory TR (ed) The Evolution of the Genome. Elsevier, San Diego, 165-221 pp.). The origin of these TEs is related to the internal degeneration of autonomous elements (Deprá et al., 2012Deprá M, Ludwig A, Valente VL and Loreto EL (2012) Mar, a MITE family of hAT transposons in Drosophila. Mobile DNA 3:1.). MITEs are distinguished from their autonomous counterparts by their high copy number, compact structure, short terminal inverted repeats, genic preference, and DNA sequence identity (Feschotte et al., 2002Feschotte C, Zhang X and Wessller SR (2002) Miniature inverted-repeat transposable elements and their relationship to established DNA transposons. In: Craig N, Craigie R, Gellert M and Lambowitz A (eds) Mobile DNA II. AMS Press, Washington, DC, pp 1147-1158., Feng, 2003Feng Y (2003) Plant MITEs: Useful tools for plant genetics and genomics. Genomics Proteomics Bioinformatics 1:90-100.).
Regarding the speciose order Coleoptera, Mariner elements have so far been described in only a few species belonging to the families Chrysomelidae, Buprestidae, Cerambycidae, Laemophloeidae, Meloidae, Scarabaeidae, Staphylinidae and Tenebrionidae (Robertson, 1993Robertson HM and Macleod EG (1993) Five major subfamilies of Mariner transposable elements in insects, including the Mediterranean fruit fly, and related arthropods. Insect Mol Biol 2:125-139.; Robertson and Macleod, 1993Robertson HM (1993) The Mariner transposable element is widespread in insects. Nature 362:241-245.; Robertson et al., 2002Robertson HM, Soto-Adames FN, Walden KK, Avancini RM and Lampe D (2002) The Mariner transposons of animals: Horizontally jumping genes. In: Kado CI (ed) Horizontal gene transfer. Academic Press, San Diego, CA, pp 173-185.; Lampe et al., 2003Lampe DJ, Witherspoon DJ, Soto-Adames FN and Robertson HM (2003) Recent horizontal transfer of mellifera subfamily Mariner transposons into insect lineages representing four different orders shows that selection acts only during horizontal transfer. Mol Biol Evol 20:554-562.; Richards et al., 2008Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, Gibbs R, Beeman RW, Brown SJ, Bucher G, et al. (2008) The genome of the model beetle and pest Tribolium castaneum. Nature 452:949-955.; Rivera-Vega and Mittapalli, 2010Rivera-Vega L and Mittapalli O (2010) Molecular characterization of Mariner-like elements in emerald ash borer, Agrilus planipennis (Coleoptera, Polyphaga). Arch Insect Biochem Physiol 74:205-216.; Oliveira et al., 2013Oliveira SG, Cabral-de-Mello DC, Moura RC and Martins C (2013) Chromosomal organization and evolutionary history of Mariner transposable elements in Scarabaeinae coleopterans. Mol Cytogenet 6:1-9.; Xavier et al., 2014Xavier C, Cabral-de-Mello DC and Moura RC (2014) Heterochromatin and molecular characterization of DsmarMITE transposable element in the beetle Dichotomius schiffleri (Coleoptera: Scarabaeidae). Genetica 142:575-581.). However, data from chromosome mapping of Mariner TEs in Coleoptera are limited to two species of Coprophanaeus, one of Diabroctis (Oliveira et al., 2013Oliveira SG, Cabral-de-Mello DC, Moura RC and Martins C (2013) Chromosomal organization and evolutionary history of Mariner transposable elements in Scarabaeinae coleopterans. Mol Cytogenet 6:1-9.) and one Dichotomius (Xavier et al., 2014Xavier C, Cabral-de-Mello DC and Moura RC (2014) Heterochromatin and molecular characterization of DsmarMITE transposable element in the beetle Dichotomius schiffleri (Coleoptera: Scarabaeidae). Genetica 142:575-581.), all genera belonging to the family Scarabaeidae. Despite the small number of studies, TEs have been associated with important evolutionary processes in Scarabeidae, such as chromosome rearrangements (Oliveira et al., 2013Oliveira SG, Cabral-de-Mello DC, Moura RC and Martins C (2013) Chromosomal organization and evolutionary history of Mariner transposable elements in Scarabaeinae coleopterans. Mol Cytogenet 6:1-9.), dispersion of 18S rDNA sites (Cabral-de-Mello et al., 2011aCabral-de-Mello DC, Moura RC and Martins C (2011a) Cytogenetic mapping of rRNAs and histone H3 genes in 14 species of Dichotomius (Coleoptera, Scarabaeidae, Scarabaeinae) beetles. Cytogenet Genome Res 134:127-135.,bCabral-de-Mello DC, Oliveira SG, Moura RC and Martins C. (2011b) Chromosomal organization of the 18S and 5S rRNAs and histone H3 genes in Scarabaeinae coleopterans: Insights into the evolutionary dynamics of multigene families and heterochromatin. BMC Genetics 12:1.), and dynamics of the repetitive DNA fraction that composes the constitutive heterochromatin (CH) in the genomes of Dichotomius species (Cabral-de-Mello et al., 2011cCabral-de-Mello DC, Moura RC, Melo AS and Martins C (2011c). Evolutionary dynamics of heterochromatin in the genome of Dichotomius beetles based on chromosomal analysis. Genetica 139:315-325.).
Cytogenetic studies have been carried out in only 18 of the 165 described Dichotomius species, including molecular cytogenetics studies in 15 species (Cabral-de-Mello et al., 2008Cabral-de-Mello DC, Oliveira SG, Ramos IC and Moura RC (2008) Karyotype differentiation patterns in species of the subfamily Scarabaeinae (Scarabaeidae, Coleoptera). Micron 38:1243-1250., 2011aCabral-de-Mello DC, Moura RC and Martins C (2011a) Cytogenetic mapping of rRNAs and histone H3 genes in 14 species of Dichotomius (Coleoptera, Scarabaeidae, Scarabaeinae) beetles. Cytogenet Genome Res 134:127-135.,b; Silva et al., 2009Silva GM, Bione EG, Cabral-de-Mello DC, Moura RC, Simões ZLP and Souza MJ (2009) Comparative cytogenetics of three species of Dichotomius (Coleoptera, Scarabaeidae). Genet Mol Biol 32:276-280.; Korasaki et al., 2012Korasaki V, Vaz-de-Mello FZ, Braga RF, Zanetti R and Louzada J (2012) Taxocoenosis of the Scarabaeinae (Coleoptera: Scarabaeidae) in Benjamin Constant, AM. Acta Amazon 42:423-432.; Xavier et al., 2014Xavier C, Cabral-de-Mello DC and Moura RC (2014) Heterochromatin and molecular characterization of DsmarMITE transposable element in the beetle Dichotomius schiffleri (Coleoptera: Scarabaeidae). Genetica 142:575-581.). This genus presents groups of closely related species (Sarmiento-Garcés and Amat-Garcia, 2009Sarmiento-Garcés R and Amat-García G (2009) Escarabajos del género Dichotomius Hope 1838 (Scarabaeidae: Scarabaeinae) en la amazonía colombiana. Rev Acad Colomb Cienc 33:285-296.), including Dichotomius (Luederwaldtinia) sericeus complex (Coleoptera: Scarabaeidae). This complex was recently taxonomically revised by Valois et al. (2017)Valois MC, Vaz-de-Mello FZ and Silva FBA (2017) Taxonomic revision of the Dichotomius sericeus (Harold, 1867) species group (Coleoptera: Scarabaeidae: Scarabaeinae). Zootaxa 4277:503–530., raising the number of species from five to eight. More specifically, D. sericeus var. aterrimus (Luederwaldt, 1929) was synonymized with D. sericeus and four new taxa, D. guaribensis, D. gilletti, D. iannuzziae, and D. catimbau have been described.
Species of the genus Dichotomius present the derived karyotype 2n = 18, Xyp, with meta-submetacentric chromosome morphology and presence of a large metacentric pair (Silva et al., 2009Silva GM, Bione EG, Cabral-de-Mello DC, Moura RC, Simões ZLP and Souza MJ (2009) Comparative cytogenetics of three species of Dichotomius (Coleoptera, Scarabaeidae). Genet Mol Biol 32:276-280.; Cabral-de-Mello et al., 2011aCabral-de-Mello DC, Moura RC and Martins C (2011a) Cytogenetic mapping of rRNAs and histone H3 genes in 14 species of Dichotomius (Coleoptera, Scarabaeidae, Scarabaeinae) beetles. Cytogenet Genome Res 134:127-135.). The constitutive heterochromatin, located in pericentromeric regions of all autosomes, show similar patterns of highly and moderately repeated DNAs (C0t-1 DNA fraction) distribution in the six analyzed species (Cabral-de-Mello et al., 2011cCabral-de-Mello DC, Moura RC, Melo AS and Martins C (2011c). Evolutionary dynamics of heterochromatin in the genome of Dichotomius beetles based on chromosomal analysis. Genetica 139:315-325.). Furthermore, the 45S rDNA is predominantly located in the distal region of the third autosome pair, whereas the 5S rDNA and H3 histone were co-located in the proximal region of the second pair in 14 analyzed species (Cabral-de-Mello et al., 2011aCabral-de-Mello DC, Moura RC and Martins C (2011a) Cytogenetic mapping of rRNAs and histone H3 genes in 14 species of Dichotomius (Coleoptera, Scarabaeidae, Scarabaeinae) beetles. Cytogenet Genome Res 134:127-135.,bCabral-de-Mello DC, Oliveira SG, Moura RC and Martins C. (2011b) Chromosomal organization of the 18S and 5S rRNAs and histone H3 genes in Scarabaeinae coleopterans: Insights into the evolutionary dynamics of multigene families and heterochromatin. BMC Genetics 12:1.).
The aim of this study was to access whether distinct populations present differential patterns of location and accumulation of Mariner-like elements. Therefore, we characterized and mapped DgmarMITE sequences in chromosomes of phylogenetically related species of the Dichotomius (Luederwaldtinia) sericeus complex belonging to different populations.
Material and Methods
Specimens sampling
All species investigated herein belong to Dichotomius (L.) sericeus complex. Dichotomius (Luederwaldtinia) gilletti and D. (L.) iannuzziae were collected in Aldeia (7º53’48’ S, 35º10’’47’W) and Igarassu (7°48’37’’S; 34°57’25’’W), remnants of the Atlantic Rain Forest in the state of Pernambuco, Brazil. Additionally, individuals of D. (L.) schiffleri and D. (L.) guaribensis were collected in Maracaípe (8°31’26”S 35°1’31”W), Pernambuco. D. (L.) guaribensis was also collected in REBIO Guaribas (6°42’41”S 35°11’17”W), Paraíba, Brazil. The individuals were collected using pitfall traps, in compliance with IBAMA/SISBIO guidelines (Permanent license No. 16278-1 for the collection of zoological material, authorization No. 41761-4 for collection in a Federal Conservation Unit for scientific purposes, and the license No. 50438-1, specific for D. (L.) schiffleri). The specimens were identified by the taxonomist Dr. Fernando Silva, from the Universidade Federal do Pará, in Brazil.
DNA extraction and isolation of the transposable element
DNA samples of the four species of Dichotomius mentioned above were obtained from the pronotum tissue. Genomic DNA was extracted according to the protocol described by Sambrook and Russell (2001)Sambrook J and Russell DW (2001) Molecular Cloning. Cold Spring Harbor Laboratory Press, New York, 2100 p.. Mariner elements were amplified by PCR using the MOS_N679 primer from Drosophila (5’GCCATATGTCGAGTT TCGTGCCA) (Zhang et al., 2001Zhang L, Dawson A and Finnegan DJ (2001) DNA-binding activity and subunit interaction of the mariner transposase. Nucleic Acids Res 29:3566-3575.).
The volume of each PCR assay was 25 μL containing 12 ng genomic DNA, 1x PCR buffer, 5 mM MgCl2, 0.2 mM dNTP (Invitrogen), 1 pmol primer, and 1 U Taq polymerase (Invitrogen). The PCR conditions were 94 ºC for 5 min, followed by 30 cycles at 94 ºC for 30 s, 49 ºC for 30 s and 72 ºC for 1.20 s, and a final extension step at 72 ºC for 5 min.
PCR products were separated by electrophoresis on 1% agarose gel. A band of approximately 500 bp obtained from D. (L.) gilletti (Supplementary Figure S1) was isolated from the gel using the Zymoclean™ Gel DNA Recovery Kit (Sinapse) according to the protocol of the manufacturer.
Cloning and sequencing
The isolated DNA fragment was cloned using the pGEM-T Easy Vector (Promega) according to manufacturer’s instructions. The insert was isolated by PCR using the M13 primer (M13F 5’-GTAAAACGACGGC CAG/M13R 5’-CAGGAAACAGCATATGAC). Concentrations of PCR reagents were the same as those described above. The PCR conditions were 95 ºC for 3 min, followed by 30 cycles at 95 ºC for 30 s, 55 ºC for 1 min and 72 ºC for 2 min, and a final extension step at 72 ºC for 5 min. For sequencing, the M13 PCR product was purified with ExoSAP-IT (Affymetrix/USB) and sequenced in an ABI3730XL automated sequencer (Applied Biosystems) by Macrogen Inc.
Editing and analysis of the transposable element
The chromatograms of forward and reverse strands of the M13 PCR product were analyzed with the Pregap4 software of the Staden package (Bonfield et al., 1995Bonfield JK, Smith KF and Staden R (1995) A new DNA sequence assembly program. Nucleic Acids Res 24:4992-4999.) in order to generate consensus sequences. Only bases with a Phred value of 20 or higher were considered in this analysis. Vector sequences were removed using the VecScreen toolVecScreen tool, http://www.ncbi.nlm.nih.gov/VecScreen/VecScreen.html (June 5, 2017).
http://www.ncbi.nlm.nih.gov/VecScreen/Ve...
. The sequence obtained (accession number: KX787885) was used as a query in GenBank BlastGenBank Blast, https://blast.ncbi.nlm.nih.gov/Blast.cgi (June 6, 2017).
https://blast.ncbi.nlm.nih.gov/Blast.cgi...
and RepBase CensorRepBase Censor, http://www.girinst.org/censor/index.php (June 6, 2017).
http://www.girinst.org/censor/index.php...
for correct identification and classification of the elements. In addition, the presence of ORFs was investigated using the ORFfinder toolORFfinder tool, http://www.ncbi.nlm.nih.gov/orffinder (June 5, 2017).
http://www.ncbi.nlm.nih.gov/orffinder...
.
Chromosome preparations, C-banding and fluorescent in situ hybridization (FISH)
Cytological preparations of our four target species were obtained by the classical testicular follicles squashing technique in 50% acetic acid. Two male individuals of each species were analyzed. C-banding was performed on D. (L.) gilletti and D. (L.) guaribensis karyotypes following Sumner (1972)Sumner AT (1972) A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res 75:304–306.. FISH was performed according to the protocol of Pinkel et al. (1986)Pinkel D, Straume T and Gray JW (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci U S A 83:2934-2938., with modifications as proposed by Cabral-de-Mello et al. (2010)Cabral-de-Mello DC, Moura RC and Martins C (2010) Chromosomal mapping of repetitive DNAs in the beetle Dichotomius geminatus provides the first evidence for an association of 5S rRNA and histone H3 genes in insects, and repetitive DNA similarity between the B chromosome and A complement. Heredity 104:393-400.. The probe of the transposable element was labeled with dUTP-digoxigenin (Roche) and detected with anti-digoxigenin-rhodamine (Roche).
Photodocumentation
Hybridization images were captured with a Leica DM 2500 epifluorescence microscope. Brightness and contrast of the images were optimized using the Photoshop CS5 program.
Results
The presence of fragments amplified by the MOS_N679 primer of Mariner elements (Figure S1) and hybridization signals of the DgmarMITE were observed in only two out of the four analyzed species, namely Dichotomius (L.) gilletti and D. (L.) guaribensis. The element isolated from D. (L.) gilletti was 496 bp-long, rich in AT (57%), and had perfect TIRs of 24 bp. The consensus sequence used as a query sequence in GenBank and RepBase revealed 100% similarity with TIRs of the AfMar2 Mariner-like element of the grasshopper Abracris flavolineata (Figure 1) (accession number: KJ829354.1). The sequence between TIRs had no similarities to previously described elements. In addition, the largest identified ORF contained only 30 amino acids and showed no similarity to any transposase.
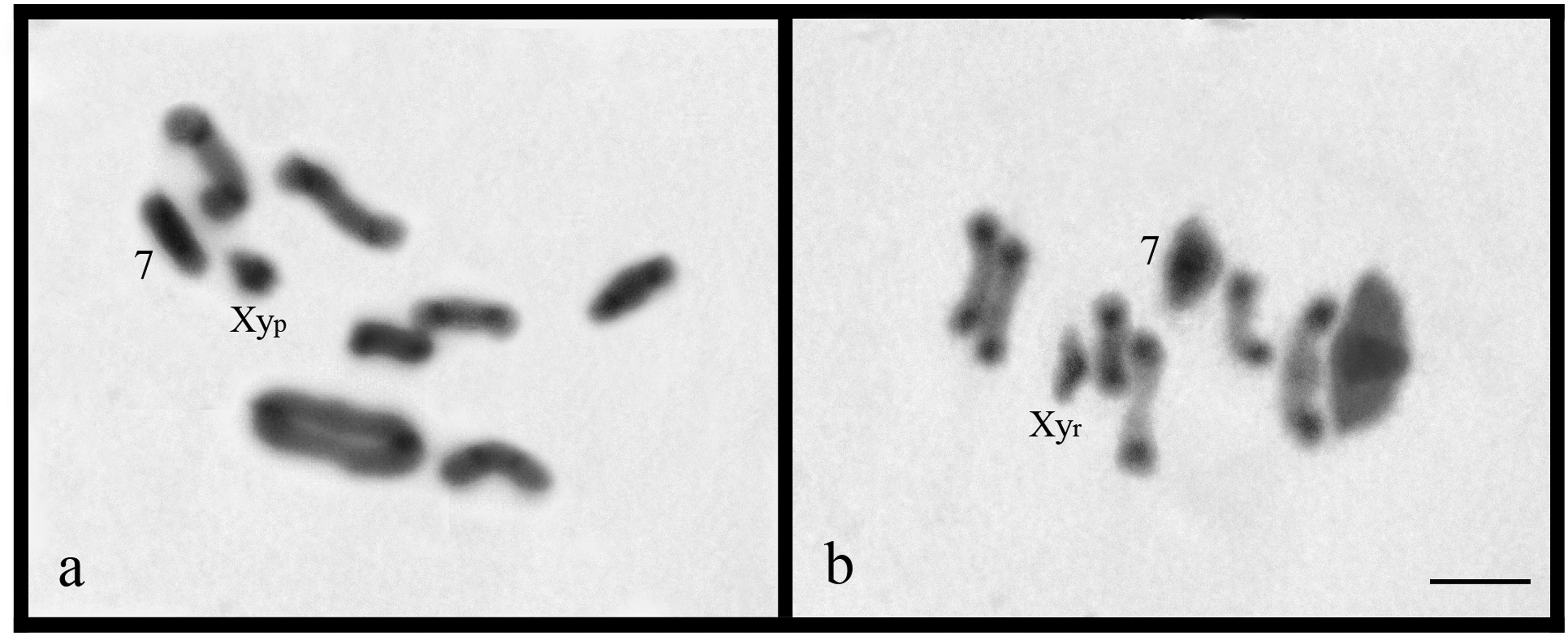
The species Dichotomius (L.) guaribensis, D. (L.) gilletti, D. (L.) iannuzziae and D. (L.) schiffleri presented similar karyotypes with 2n = 18, and meta-submetacentric chromosomal morphology. However, distinct sexual determination systems were obverved: D. (L.) gilletti, and D. (L.) iannuzziae had a Xyp system, whereas D. (L.) schiffleri and D. (L.) guaribensis presented a Xyr sex bivalent configuration (Figure 2). C-banding revealed pericentromeric constitutive heterochromatin in all autosomes, and additionally, along the entire length of the seventh bivalent and X chromosome of Dichotomius (L.) gilletti and D. (L.) guaribensis (Figure 2a,b).
C-banding in metaphase I of Dichotomius (L.) gilletti (a) and metaphase I of D. (L.) guaribensis (b). Bar = 5 μm.
Mapping of DgmarMITE probes on the karyotype of D. (L.) gilletti revealed signals in all chromosomes, except for pairs five and seven of the Igarassu population (Figure 3a), and pair five of the Aldeia population (Figure 3b). Overall, DgmarMITE sequences were predominantly located in euchromatic regions in individuals from both populations, except in the Igarassu population, for which signals were detected at heterochromatic regions of chromosome pairs six and eight (Figure 3a). Similarly, in Aldeia population, DgmarMITE was restricted to the heterochromatic region of pair two (Figure 3b). In addition, five heteromorphic pairs were observed in Aldeia individuals (Figure 3b).
Fluorescent in situ hybridization of the element DgmarMITE in meiotic cells of two individuals of Dichotomius (Luederwaldtinia) gilletti (a, b) and D. (L.) guaribensis (c, d). (a) Metaphase I of an individual from Igarassu population; (b) diplotene of specimen from Aldeia population; (c) metaphase I of individual from Guaribas population; (d) metaphase II of specimen from Maracaípe population. Note the localization and the size of the signals between populations. Bar = 5 μm.
Mapping of DgmarMITE probes on the karyotype of D. (L.) guaribensis revealed their location in heterochromatic regions of all autosomes and of the X chromosome in both populations (Figure 3c,d). Additional signal was observed on the y chromosome of the Guaribas population (Figure 3c). Furthermore, DgmarMITE sites were observed in euchromatic regions of all autosomes, except pair eight, in specimens from Maracaípe (Figure 3d). Overall, stronger FISH signals were observed in the karyotypes of individuals from Guaribas when compared to specimens from Maracaípe (Figure 3c,d).
Discussion
The karyotype observed in D. (L.) guaribensis (2n = 18) is considered derived from the ancestral number reported for the family Scarabaeidae (2n = 20), but conserved in most species of Dichotomius. The configuration of the sexual bivalent (Xyr), which has been reported so far only in D. schiffleri, also differs from the ancestral Scarabeidae Xyp (Cabral-de-Mello et al., 2008Cabral-de-Mello DC, Oliveira SG, Ramos IC and Moura RC (2008) Karyotype differentiation patterns in species of the subfamily Scarabaeinae (Scarabaeidae, Coleoptera). Micron 38:1243-1250., 2011aCabral-de-Mello DC, Moura RC and Martins C (2011a) Cytogenetic mapping of rRNAs and histone H3 genes in 14 species of Dichotomius (Coleoptera, Scarabaeidae, Scarabaeinae) beetles. Cytogenet Genome Res 134:127-135.; Silva et al., 2009Silva GM, Bione EG, Cabral-de-Mello DC, Moura RC, Simões ZLP and Souza MJ (2009) Comparative cytogenetics of three species of Dichotomius (Coleoptera, Scarabaeidae). Genet Mol Biol 32:276-280.; Xavier et al., 2014Xavier C, Cabral-de-Mello DC and Moura RC (2014) Heterochromatin and molecular characterization of DsmarMITE transposable element in the beetle Dichotomius schiffleri (Coleoptera: Scarabaeidae). Genetica 142:575-581.). The derived karyotypes of D. gilletti and D. iannuzziae observed in this study are similar to those described by Cabral-de Mello et al. (2011a)Cabral-de-Mello DC, Moura RC and Martins C (2011a) Cytogenetic mapping of rRNAs and histone H3 genes in 14 species of Dichotomius (Coleoptera, Scarabaeidae, Scarabaeinae) beetles. Cytogenet Genome Res 134:127-135. prior to the taxonomic revision by Valois et al. (2017)Valois MC, Vaz-de-Mello FZ and Silva FBA (2017) Taxonomic revision of the Dichotomius sericeus (Harold, 1867) species group (Coleoptera: Scarabaeidae: Scarabaeinae). Zootaxa 4277:503–530.. In Cabral-de Mello et al. (2011a)Cabral-de-Mello DC, Moura RC and Martins C (2011a) Cytogenetic mapping of rRNAs and histone H3 genes in 14 species of Dichotomius (Coleoptera, Scarabaeidae, Scarabaeinae) beetles. Cytogenet Genome Res 134:127-135., these species were referred to as D. sericeus and D. laevicollis, respectively. The presence of constitutive heterochromatic blocks in pericentromeric regions of all autosomes, as observed in D. (L.) gilletti and D. (L.) guaribensis, is a common feature in the genus Dichotomius (Cabral-de-Mello et al., 2011cCabral-de-Mello DC, Moura RC, Melo AS and Martins C (2011c). Evolutionary dynamics of heterochromatin in the genome of Dichotomius beetles based on chromosomal analysis. Genetica 139:315-325.), and has also been reported for the other two species investigated herein: D. (L.) iannuzziae (Cabral-de-Mello et al., 2011cCabral-de-Mello DC, Moura RC, Melo AS and Martins C (2011c). Evolutionary dynamics of heterochromatin in the genome of Dichotomius beetles based on chromosomal analysis. Genetica 139:315-325.) and D. (L.) schiffleri (Xavier et al., 2014Xavier C, Cabral-de-Mello DC and Moura RC (2014) Heterochromatin and molecular characterization of DsmarMITE transposable element in the beetle Dichotomius schiffleri (Coleoptera: Scarabaeidae). Genetica 142:575-581.).
DgmarMITE, the TE characterized here, presented features shared by all MITEs such as a large copy number, which was observable with FISH resolution, lack of transposase coding, AT richness, and conservation of TIR structure (Kuang et al., 2009Kuang H, Padmanabhan C, Li F, Kamei A, Bhaskar PB, Ouyang S, Jiang J, Buell CR and Baker B (2009) Identification of miniature inverted-repeat transposable elements (MITEs) and biogenesis of their siRNAs in the Solanaceae: New functional implications for MITEs. Genome Res 19:42-56.). In most cases, sequence similarity between a MITE and its closest element is restricted to the TIRs (Feschotte and Pritham, 2007Feschotte C and Pritham EJ (2007) DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet 41:331-368.). The similarity between the TIRs of DgmarMITE and the AfMar2 element of A. flavolineata indicates that the former belongs to the Mariner family.
The presence of DgmarMITE in the taxonomically similar species D. (L.) gilletti and D. (L.) guaribensis [considering the genus revision of Valois et al. (2017)Valois MC, Vaz-de-Mello FZ and Silva FBA (2017) Taxonomic revision of the Dichotomius sericeus (Harold, 1867) species group (Coleoptera: Scarabaeidae: Scarabaeinae). Zootaxa 4277:503–530.] and therefore possibly phylogenetically closer, suggests an origin of this element in the common ancestor of these species. An alternative hypothesis is that this element may have originated independently in the species D. (L.) gilletti and D. (L.) guaribensis by horizontal transfer events, which are frequently observed for Mariner-like elements (Robertson, 1995Robertson HM (1995) The Tcl-Mariner superfamily of transposons in animals. J Insect Physiol 41:99-105.; Robertson and Lampe, 1995Robertson HM (1995) The Tcl-Mariner superfamily of transposons in animals. J Insect Physiol 41:99-105.), including MITEs, as described previously for the Stowaway element in the plant family Pooideae (Minaya et al., 2013Minaya M, Pimentel M, Mason-Gamer R and Catalan P (2013) Distribution and evolutionary dynamics of Stowaway Miniature Inverted repeat Transposable Elements (MITEs) in grasses. Mol Phylogenet Evol 68:106-118.). The DgmarMITE origin may be recent or not however, since older TEs accumulate preferentially in heterochromatic regions (Junakovic et al., 1998Junakovic N, Terrinoni A, Di Franco C, Vieira C and Loevenbruck C (1998) Accumulation of transposable elements in the heterochromatin and on the Y chromosome of Drosophila simulans and Drosophila melanogaster. J Mol Evol 46:661-668.), as has been previously proposed for other TEs in Scarabaeidae (Oliveira et al., 2013Oliveira SG, Cabral-de-Mello DC, Moura RC and Martins C (2013) Chromosomal organization and evolutionary history of Mariner transposable elements in Scarabaeinae coleopterans. Mol Cytogenet 6:1-9.). Genome colonization of this MITE possibly occurred earlier in D. (L.) guaribensis than in D. (L.) gilletti. In D. (L.) gilletti, the preferential location in euchromatic regions, the presence of heteromorphic pairs in the population of Aldeia, and absence of the signal in one or two chromosome pairs, suggest that this element originated most recently.
In addition to D. (L.) gilletti, predominantly euchromatic signals have been reported for TEs in D. (L.) schiffleri (Xavier et al., 2014Xavier C, Cabral-de-Mello DC and Moura RC (2014) Heterochromatin and molecular characterization of DsmarMITE transposable element in the beetle Dichotomius schiffleri (Coleoptera: Scarabaeidae). Genetica 142:575-581.), for the grasshopper species Eyprepocnemis plorans (Montiel et al., 2012Montiel EE, Cabrero J, Camacho JPM and López-León DM (2012) Gypsy, RTE and Mariner transposable elements populate Eyprepocnemis plorans genome. Genetica 140:365-374.) and A. flavolineata (Palacios-Gimenez et al., 2014Palacios-Gimenez OM, Bueno D and Cabral-de-Mello DC (2014) Chromosomal mapping of two Mariner-like elements in the grasshopper Abracris flavolineata (Orthoptera: Acrididae) reveals enrichment in euchromatin. Eur J Entomol 111:329-334.). The occurrence of DgmarMITE in euchromatic regions can influence gene expression and/or gene and chromosome mutations (Kidwell and Lish, 2000Kidwell MG and Lisch D (2000) Transposable elements and host genome evolution. Trends Ecol Evol 15:95-99.; Feschotte and Pritham, 2007Feschotte C and Pritham EJ (2007) DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet 41:331-368.). However, it is also possible that the element is inserted in pseudogenes or even other dispersed repetitive sequences in euchromatin, as proposed for Mariner family elements of E. plorans (Montiel et al., 2012Montiel EE, Cabrero J, Camacho JPM and López-León DM (2012) Gypsy, RTE and Mariner transposable elements populate Eyprepocnemis plorans genome. Genetica 140:365-374.). On the other hand, the presence of MITEs in heterochromatic regions, as observed for DgmarMITE in D. (L.) guaribensis, is not common, since these TEs are preferentially associated with genes (Lu et al., 2011Lu C, Chen J, Zhang Y, Hu Q, Su W and Kuang H (2011) Miniature inverted–repeat transposable elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol Biol Evol 29:1005-1017.). However, heterochromatic enrichment of these elements has been described in other organisms, such as in the insect Anopheles gambiae (Quesneville et al., 2006Quesneville H, Nouaud D and Anxolabéhère D (2006) P elements and MITE relatives in the whole genome sequence of Anopheles gambiae. BMC Genomics 7:214.) and in the plants Oryza sativa (Lu et al., 2011Lu C, Chen J, Zhang Y, Hu Q, Su W and Kuang H (2011) Miniature inverted–repeat transposable elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol Biol Evol 29:1005-1017.) and Arabidopsis thaliana (Guo et al., 2017Guo C, Spinelli M, Ye C, Li QQ and Liang C (2017) Genome-wide comparative analysis of miniature inverted repeat transposable elements in 19 Arabidopsis thaliana ecotype accessions. Sci Rep 7:2634.).
Mapping of DgmarMITE in D. (L.) gilletti and D. (L.) guaribensis showed variation in the location and number of sites between species and populations. These findings suggest that this non-autonomous element may be cross-mobilized to different regions of host genomes using the transposase of either an older or a newly emerged transposon, in this latter case DgmarMITE accumulation occurs by a process known as “snowball effect” (Feschotte et al., 2005Feschotte C, Osterlund MT, Peeler R and Wessler SR (2005) DNA-binding specificity of rice Mariner-like transposases and interactions with Stowaway MITEs. Nucleic Acids Res 33:2153-2165.). The transposase used by DgmarMITE could belong to a, closely related TE as observed between the inactive peach element and the transposase of Mariner-like Mos1 in Drosophila melanogaster (Garza et al., 1991Garza D, Medhora M, Koga A and Hartl DL (1991) Introduction of the transposable element Mariner into the germline of Drosophila melanogaster. Genetics 128:303-310.), or to a phylogenetically distant TE, as observed between an element of the Stowaway family and Osmar transposase, an autonomous Mariner-like element in the rice genome (Feschotte et al., 2003Feschotte C, Swamy L and Wessler SR (2003) Genome-wide analysis of Mariner-like transposable elements in rice reveals complex relationships with Stowaway miniature inverted repeat transposable elements (MITEs). Genetics 163:747-758.; Yang et al., 2009Yang G, Nagel DH, Feschotte C, Hancock CN and Wessler SR (2009) Tuned for transposition: molecular determinants underlying the hyperactivity of a Stowaway MITE. Science 325:1391-1394.).
With respect to copy-number variation in heterochromatic regions of D. (L.) guaribensis chromosomes, an increase in DgmarMITE copy number in the Guaribas population probably results from transposition-independent events, including ectopic recombination and concerted evolution. The latter has been observed for highly repetitive DNA sequences such as Mariner elements found in the heterochromatin of Drosophila erecta (Lohe et al., 1995Lohe AR, Moriyama EN, Lidholm DA and Hartl DL (1995) Horizontal transmission, vertical inactivation, and stochastic loss of Mariner-like transposable elements. Mol Biol Evol 12:62-72.). An alternative hypothesis to explain this variation is that this element is undergoing a reverse process with quantitative and random copy loss in the genomes of individuals from Maracaípe population. In this scenario, DgmarMITE would be undergoing senescence, the last stage of the transposable element “life cycle”, as described by Kidwell and Lisch (2000)Kidwell MG and Lisch D (2000) Transposable elements and host genome evolution. Trends Ecol Evol 15:95-99..
Mapping of DgmarMITE in species of the Dichotomius (Luederwaldtinia) sericeus complex contributed to increase our knowledge about the location and distribution of TEs in dung beetle genomes. This analysis also revealed the accumulation of DgmarMITE in the karyotype of two species. Plausible mechanisms underlying such accumulation include the occurrence of cross-mobilization and/or ectopic recombination in heterochromatic regions. However, we cannot completely rule out the possible involvement of other molecular mechanisms discussed here. Therefore, further characterization and chromosome mapping should be extended to other species within this complex of species.
Acknowledgments
We would like to thank Dr. Silva, F.B.A. (UFPA) for her assistance in identifying the specimens. In addition, we would like to thank Dra. Oliveira SG, for her suggestions on drafts of this manuscript. The study was funded by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) through doctoral scholarships to ICA and CX; Funds from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) covered a scientific initiation scholarship to RGCC and research grant to RCM (PQ2 CNPQ- 305298/2014-3). Support was also obtained from the Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco (FACEPE APQ - 0777-2.02/15).
References
- Bonfield JK, Smith KF and Staden R (1995) A new DNA sequence assembly program. Nucleic Acids Res 24:4992-4999.
- Cabral-de-Mello DC, Oliveira SG, Ramos IC and Moura RC (2008) Karyotype differentiation patterns in species of the subfamily Scarabaeinae (Scarabaeidae, Coleoptera). Micron 38:1243-1250.
- Cabral-de-Mello DC, Moura RC and Martins C (2010) Chromosomal mapping of repetitive DNAs in the beetle Dichotomius geminatus provides the first evidence for an association of 5S rRNA and histone H3 genes in insects, and repetitive DNA similarity between the B chromosome and A complement. Heredity 104:393-400.
- Cabral-de-Mello DC, Moura RC and Martins C (2011a) Cytogenetic mapping of rRNAs and histone H3 genes in 14 species of Dichotomius (Coleoptera, Scarabaeidae, Scarabaeinae) beetles. Cytogenet Genome Res 134:127-135.
- Cabral-de-Mello DC, Oliveira SG, Moura RC and Martins C. (2011b) Chromosomal organization of the 18S and 5S rRNAs and histone H3 genes in Scarabaeinae coleopterans: Insights into the evolutionary dynamics of multigene families and heterochromatin. BMC Genetics 12:1.
- Cabral-de-Mello DC, Moura RC, Melo AS and Martins C (2011c). Evolutionary dynamics of heterochromatin in the genome of Dichotomius beetles based on chromosomal analysis. Genetica 139:315-325.
- Deprá M, Ludwig A, Valente VL and Loreto EL (2012) Mar, a MITE family of hAT transposons in Drosophila Mobile DNA 3:1.
- Feng Y (2003) Plant MITEs: Useful tools for plant genetics and genomics. Genomics Proteomics Bioinformatics 1:90-100.
- Feschotte C and Pritham EJ (2007) DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet 41:331-368.
- Feschotte C, Zhang X and Wessller SR (2002) Miniature inverted-repeat transposable elements and their relationship to established DNA transposons. In: Craig N, Craigie R, Gellert M and Lambowitz A (eds) Mobile DNA II. AMS Press, Washington, DC, pp 1147-1158.
- Feschotte C, Swamy L and Wessler SR (2003) Genome-wide analysis of Mariner-like transposable elements in rice reveals complex relationships with Stowaway miniature inverted repeat transposable elements (MITEs). Genetics 163:747-758.
- Feschotte C, Osterlund MT, Peeler R and Wessler SR (2005) DNA-binding specificity of rice Mariner-like transposases and interactions with Stowaway MITEs. Nucleic Acids Res 33:2153-2165.
- Garza D, Medhora M, Koga A and Hartl DL (1991) Introduction of the transposable element Mariner into the germline of Drosophila melanogaster Genetics 128:303-310.
- Guo C, Spinelli M, Ye C, Li QQ and Liang C (2017) Genome-wide comparative analysis of miniature inverted repeat transposable elements in 19 Arabidopsis thaliana ecotype accessions. Sci Rep 7:2634.
- Junakovic N, Terrinoni A, Di Franco C, Vieira C and Loevenbruck C (1998) Accumulation of transposable elements in the heterochromatin and on the Y chromosome of Drosophila simulans and Drosophila melanogaster J Mol Evol 46:661-668.
- Kidwell MG (2005) Transposable Elements. In: Gregory TR (ed) The Evolution of the Genome. Elsevier, San Diego, 165-221 pp.
- Kidwell MG and Lisch D (2000) Transposable elements and host genome evolution. Trends Ecol Evol 15:95-99.
- Korasaki V, Vaz-de-Mello FZ, Braga RF, Zanetti R and Louzada J (2012) Taxocoenosis of the Scarabaeinae (Coleoptera: Scarabaeidae) in Benjamin Constant, AM. Acta Amazon 42:423-432.
- Kuang H, Padmanabhan C, Li F, Kamei A, Bhaskar PB, Ouyang S, Jiang J, Buell CR and Baker B (2009) Identification of miniature inverted-repeat transposable elements (MITEs) and biogenesis of their siRNAs in the Solanaceae: New functional implications for MITEs. Genome Res 19:42-56.
- Lampe DJ, Churchill MEA and Robertson HM (1996) A purified Mariner transposase is sufficient to mediate transposition in vitro EMBO J 15:5470-5479.
- Lampe DJ, Witherspoon DJ, Soto-Adames FN and Robertson HM (2003) Recent horizontal transfer of mellifera subfamily Mariner transposons into insect lineages representing four different orders shows that selection acts only during horizontal transfer. Mol Biol Evol 20:554-562.
- Lohe AR, Moriyama EN, Lidholm DA and Hartl DL (1995) Horizontal transmission, vertical inactivation, and stochastic loss of Mariner-like transposable elements. Mol Biol Evol 12:62-72.
- Lu C, Chen J, Zhang Y, Hu Q, Su W and Kuang H (2011) Miniature inverted–repeat transposable elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa Mol Biol Evol 29:1005-1017.
- Minaya M, Pimentel M, Mason-Gamer R and Catalan P (2013) Distribution and evolutionary dynamics of Stowaway Miniature Inverted repeat Transposable Elements (MITEs) in grasses. Mol Phylogenet Evol 68:106-118.
- Montiel EE, Cabrero J, Camacho JPM and López-León DM (2012) Gypsy, RTE and Mariner transposable elements populate Eyprepocnemis plorans genome. Genetica 140:365-374.
- Oliveira SG, Bao W, Martins C and Jurka J (2012) Horizontal transfers of Mariner transposons between mammals and insects. Mobile DNA 3:1-6.
- Oliveira SG, Cabral-de-Mello DC, Moura RC and Martins C (2013) Chromosomal organization and evolutionary history of Mariner transposable elements in Scarabaeinae coleopterans. Mol Cytogenet 6:1-9.
- Palacios-Gimenez OM, Bueno D and Cabral-de-Mello DC (2014) Chromosomal mapping of two Mariner-like elements in the grasshopper Abracris flavolineata (Orthoptera: Acrididae) reveals enrichment in euchromatin. Eur J Entomol 111:329-334.
- Pinkel D, Straume T and Gray JW (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci U S A 83:2934-2938.
- Plasterk RHA, Izsvák Z and Ivics Z (1999) Resident aliens the Tc1/Mariner superfamily of transposable elements. Trends Genet 15:326-332.
- Quesneville H, Nouaud D and Anxolabéhère D (2006) P elements and MITE relatives in the whole genome sequence of Anopheles gambiae BMC Genomics 7:214.
- Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, Gibbs R, Beeman RW, Brown SJ, Bucher G, et al. (2008) The genome of the model beetle and pest Tribolium castaneum Nature 452:949-955.
- Rivera-Vega L and Mittapalli O (2010) Molecular characterization of Mariner-like elements in emerald ash borer, Agrilus planipennis (Coleoptera, Polyphaga). Arch Insect Biochem Physiol 74:205-216.
- Robertson HM (1993) The Mariner transposable element is widespread in insects. Nature 362:241-245.
- Robertson HM (1995) The Tcl-Mariner superfamily of transposons in animals. J Insect Physiol 41:99-105.
- Robertson HM and Macleod EG (1993) Five major subfamilies of Mariner transposable elements in insects, including the Mediterranean fruit fly, and related arthropods. Insect Mol Biol 2:125-139.
- Robertson HM and Lampe D (1995) Distribution of transposable elements in Arthropods. Annu Rev Entomol 40:333-357.
- Robertson HM, Soto-Adames FN, Walden KK, Avancini RM and Lampe D (2002) The Mariner transposons of animals: Horizontally jumping genes. In: Kado CI (ed) Horizontal gene transfer. Academic Press, San Diego, CA, pp 173-185.
- Sambrook J and Russell DW (2001) Molecular Cloning. Cold Spring Harbor Laboratory Press, New York, 2100 p.
- Sarmiento-Garcés R and Amat-García G (2009) Escarabajos del género Dichotomius Hope 1838 (Scarabaeidae: Scarabaeinae) en la amazonía colombiana. Rev Acad Colomb Cienc 33:285-296.
- Silva GM, Bione EG, Cabral-de-Mello DC, Moura RC, Simões ZLP and Souza MJ (2009) Comparative cytogenetics of three species of Dichotomius (Coleoptera, Scarabaeidae). Genet Mol Biol 32:276-280.
- Sumner AT (1972) A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res 75:304–306.
- Valois MC, Vaz-de-Mello FZ and Silva FBA (2017) Taxonomic revision of the Dichotomius sericeus (Harold, 1867) species group (Coleoptera: Scarabaeidae: Scarabaeinae). Zootaxa 4277:503–530.
- Wallau GL, Capy P, Loreto E and Hua-Van A (2014) Genomic landscape and evolutionary dynamics of Mariner transposable elements within the Drosophila genus. BMC Genomics 15:727.
- Xavier C, Cabral-de-Mello DC and Moura RC (2014) Heterochromatin and molecular characterization of DsmarMITE transposable element in the beetle Dichotomius schiffleri (Coleoptera: Scarabaeidae). Genetica 142:575-581.
- Yang G, Nagel DH, Feschotte C, Hancock CN and Wessler SR (2009) Tuned for transposition: molecular determinants underlying the hyperactivity of a Stowaway MITE. Science 325:1391-1394.
- Zhang L, Dawson A and Finnegan DJ (2001) DNA-binding activity and subunit interaction of the mariner transposase. Nucleic Acids Res 29:3566-3575.
Internet Resources
- GenBank Blast, https://blast.ncbi.nlm.nih.gov/Blast.cgi (June 6, 2017).
» https://blast.ncbi.nlm.nih.gov/Blast.cgi - ORFfinder tool, http://www.ncbi.nlm.nih.gov/orffinder (June 5, 2017).
» http://www.ncbi.nlm.nih.gov/orffinder - RepBase Censor, http://www.girinst.org/censor/index.php (June 6, 2017).
» http://www.girinst.org/censor/index.php - VecScreen tool, http://www.ncbi.nlm.nih.gov/VecScreen/VecScreen.html (June 5, 2017).
» http://www.ncbi.nlm.nih.gov/VecScreen/VecScreen.html
Supplementary material
The following online material is available for this article:
-
Associate Editor: Igor Schneider
Publication Dates
-
Publication in this collection
04 June 2018 -
Date of issue
Apr./June 2018
History
-
Received
26 July 2017 -
Accepted
23 Nov 2017