Abstract
The leachate generated by the decomposition of animal carcass has been implicated as an environmental contaminant surrounding the burial site. High-throughput nucleotide sequencing was conducted to investigate the bacterial communities in leachates from the decomposition of pig carcasses. We acquired 51,230 reads from six different samples (1, 2, 3, 4, 6 and 14 week-old carcasses) and found that sequences representing the phylum Firmicutes predominated. The diversity of bacterial 16S rRNA gene sequences in the leachate was the highest at 6 weeks, in contrast to those at 2 and 14 weeks. The relative abundance of Firmicutes was reduced, while the proportion of Bacteroidetes and Proteobacteria increased from 3–6 weeks. The representation of phyla was restored after 14 weeks. However, the community structures between the samples taken at 1–2 and 14 weeks differed at the bacterial classification level. The trend in pH was similar to the changes seen in bacterial communities, indicating that the pH of the leachate could be related to the shift in the microbial community. The results indicate that the composition of bacterial communities in leachates of decomposing pig carcasses shifted continuously during the study period and might be influenced by the burial site.
pig; decomposition; leachate; bacterial community; pyrosequencing
Introduction
Foot and mouth disease (FMD) is highly contagious and is caused by members of the genus Aphthovirus, represented by various serotypes, such as A, O, C, SAT-1, SAT-2, SAT-3, and Asia-1 (Brooksby, 1982Brooksby JB (1982) Portraits of viruses: foot-and-mouth disease virus. Intervirology 18:1–23.; Seellers, 1995Seellers RF (1995) Growth and titration of the viruses of foot-and mouth disease and vesicular stomatitis in kidney monolayer tissue cultures. Nature 176:121–124.).
FMD is especially fatal in young animals as a result of myocardial damage, while the mortality rate of FMD might be low in adult animals (Sharma and Das, 1984Sharma PK and Das SK (1984) Occurrence of foot-and mouth disease and distribution of virus type in the hill states of North Eastern region of India. Indian J Anim Sci 4:117–118.; Woodbury, 1995Woodbury EL (1995) A review of the possible mechanisms for the persistence of foot-and-mouth disease virus. Epidemiol Infect 114:1–13.; Domingo et al., 1990Domingo E, Mateu MG, Martínez MA, Dopazo J, Moya A and Sobrino F (1990) Genetic variability and antigenic diversity of foot-and-mouth disease virus. In: Kurstak E, Marusyk RG, Murphy SA and VanRegenmortel MHV (eds) Applied Virology Research, vol 2: Virus Variation and Epidemiology. Plenum Publishing Corp, New York, pp 233–266.; Gruman and Baxt, 2004Gruman MJ and Baxt B (2004) Foot-and-mouth disease. Clin Microbiol Rev 17:465–493.). Therefore, FMD-infected animals are slaughtered immediately and buried according to guidance on animal carcass mass burial procedures developed by each country. Despite an extensive nationwide quarantine effort, South Korea experienced the worst FMD epidemic in the country’s history during 2010–2011, resulting in the culling of more than 3.5 million heads of Artiodactyla, including cattle, goat and pigs (Ahn, 2012Ahn HK (2012) 2010 FMD outbreak in Korea: Goverment’s response to this emergency and important lessons learned. Keynote presentation at 4th International Symposium on managing Animal Mortality, Products, By Products, and Associated Health Risk, May 21–24, Dearborn, Michigan, University of Maine.).
Although burial is the most common method used to dispose of large numbers of deceased livestock, the potential pathogenic content of the carcass associated health problems to residents living near the burial site have resulted in widespread public health concerns (NRC, 2002NRC (2002) Biosolids Applied to Land: Advancing Standards and Practices. National Research Council, Washington DC, 344 pp.). Moreover, leachates derived from the decomposition of animal carcasses consist of organic intermediates, volatile chemicals, and saprotrophic organisms, which may pose a high contamination risk to soil and groundwater when leaking through the basement layer (Christensen et al., 1994Christensen TH, Kjeldsen P, Albrechtsen HJ, Heron G, Nielsen PH, Bjerg PL and Holm PE (1994) Attenuation of landfill leachate pollutants in aquifers. Crit Rev Env Sci Tec 24:119–202.; Röling et al., 2001Röling WFM, van Breukelen BM, Braster M, Lin B and van Verseveld HW (2001) Relationships between microbial community structure and hydrochemistry in a landfill leachate-polluted aquifer. Appl Environ Microb 67:4619–4629.; Tian et al., 2005Tian Y, Yang H, Wu X and Li DT (2005) Molecular analysis of microbial community in a groundwater sample polluted by landfill leachate and seawater. J Zhejiang Univ Sci 6:165–170.).
The rate at which animal carcasses decompose in the burial pit can be affected by temperature, humidity, the depth of the pit, chemical composition, and other environmental factors (Mann et al., 1990Mann RW, Bass WM and Meadows L (1990) Time since death and decomposition of the human body: variables and observations in case and experimental field studies. J Forensic Sci 35:103–111.; McDaniel, 1991McDaniel HA (1991) Environmental protection during animal disease eradication programmes. Rev Sci Tech OIE 10:867–884.; Vass et al., 1992Vass AA, Bass WM, Wolt JD, Foss JE and Ammons JT (1992) Time since death determinations of human cadavers using soil solution. J Forensic Sci 37:1236–1253.; Paul and Clark, 1996Paul EA and Clark FE (1996) Soil Microbiology and Biochemistry. 2nd edition. Academic Press, San Diego, 340 pp.; Gill-King, 1997Gill-King H (1997) Chemical and ultrastructural aspects of decomposition. In: Haglund WD and Sorg MH (eds) Forensic Taphonomy: The Postmortem Fate of Human Remains. CRC Press, New York, pp 93–104.; Carter and Tibbett, 2006Carter DO and Tibbett M (2006) Microbial decomposition of skeletal muscle tissue (Ovis aries) in a sandy loam soil at different temperatures. Soil Biol Biochem 38:1139–1145.; Pratt et al., 2010Pratt DL, Dumonceaux TJ and Fonstad TA (2010) Determination of Microbial Communities beneath Livestock Burial Sites. DOI: 10.13031/2013.36276 Conference: ASABE/CSBE North Central Intersectional Meeting.Saskatoon, SK.
https://doi.org/10.13031/201336276...
). Generally, temperature and pH can accelerate microbial decomposition by changing the soil microhabitat (Paul and Clark, 1996Paul EA and Clark FE (1996) Soil Microbiology and Biochemistry. 2nd edition. Academic Press, San Diego, 340 pp.; Blagodatskaya and Anderson, 1998Blagodatskaya EV and Anderson TH (1998) Interactive effects of pH and substrate quality on the fungal-to-bacterial ratio and QCO (2) of microbial communities in forest soils. Soil Biol Biochem 30:1269–1274.). For example, the metabolic activity of a mesophilic microbial population increased approximately two-fold with an increase in temperature from 10 °C to 30–35 °C (Van’tHoff, 1898Van’tHoff JH (1898) Lectures on Theoretical and Physical Chemistry. Part 1. Chemical Dynamics. Edward Arnold, London, pp. 224–229.; Conant et al., 2004Conant RT, Dalla-Betta P, Klopatek CC and Klopatek JA (2004) Controls on soil respiration in semiarid soils. Soil Biol Biochem 36:945–951.). Moreover, pH has a pivotal role in the type of microorganisms that predominate in different soils (Lynch and Hobbie, 1988Lynch JM and Hobbie JE (1988) Micro-Organisms in Action: Concepts and Applications in Microbial Ecology. Blackwell Scientific Publications, Oxford, 363 pp.; Matthies et al., 1997Matthies C, Erhard HP and Drake HL (1997) Effects of pH on the comparative culturability of fungi and bacteria from acidic and less acidic forest soils. J Basic Microb 37:335–343.). Other environmental factors, including moisture, dissolved oxygen, conductivity and the like also play an important role in the microbial community composition (Keener et al., 2000Keener HM, Elwell DL and Monnin MJ (2000) Procedures and equations for sizing of structures and windrows for composting animal mortalities. Appl Eng Agric 16:681–692.; Pettersson M, (2004, Doctoral thesis, Lund University, Sweden). Soil properties may also affect the microbial community (Carter et al., 2007Carter DO, Yellowlees D and Tibbett M (2007) Cadaver decomposition in terrestrial ecosystems. Naturwissenschaften 94:12–24.) and, hence, the carcass decomposition process (Pfeiffer et al., 1998Pfeiffer S, Milne S and Stevenson RM (1998) The natural decomposition of adipocere. J Forensic Sci 43:368–370.).
In a forensic study, the bodies of obese people decomposed faster than those of average weight individuals due to the greater amount of liquid present in the tissues, which contributes to the spread and proliferation of bacteria (Campobasso et al., 2001Campobasso CP, Di Vella G and Introna F (2001) Factors affecting decomposition and Diptera colonization. Forensic Sci Int 120:18–27.). The high organic load of the leachate could cause drastic changes in aquifer geochemistry and microbiology downstream of landfills (Roling et al., 2001Röling WFM, van Breukelen BM, Braster M, Lin B and van Verseveld HW (2001) Relationships between microbial community structure and hydrochemistry in a landfill leachate-polluted aquifer. Appl Environ Microb 67:4619–4629.), and leachates originating from the decomposition of buried pig carcasses, as investigated here, could cause similar changes in soil and in the geochemical characteristics of water as well.
The methods for burying dead livestock or the characteristics of uncultured microbial communities in the burial site have not been studied in detail. The presence of many uncultured bacteria has been confirmed by polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) and phospholipid fatty acid (PLFA) profiling. However, such studies primarily focused on specific microbial communities in landfill leachate plumes (Ludvigsen et al., 1997Ludvigsen L, Albrechtsen HJ, Holst H and Christensen TH (1997) Correlating phospholipid fatty acids (PLFA) in a landfill leachate polluted aquifer with biogeochemical factors by multivariate statistical methods. FEMS Microbiol Rev 20:447–460.; Sasaki et al., 2009Sasaki H, Nonaka J, Otawa K, Kitazume O, Asano R, Sasaki T and Nakai Y (2009) Analysis of the structure of the bacterial community in the livestock manure-based composting process. Asian-Aust J Anim 22:113–118.; Yang et al., 2012Yang SH, Hong SH, Cho SB, Lim JS, Bae SE, Ahn HK and Lee EY (2012) Characterization of microbial community in the leachate associated with the decomposition of entombed pigs. J Microbiol Biotechn 22:1330–5.). The advent of next generation sequencing techniques, such as bar-coded pyrosequencing, can elucidate microbial community structures at a much higher resolution than previously possible. Moreover, the large dataset produced by pyrosequencing enables the detection of rare microbes and to perform phylogenetic comparisons of microbes living in a particular environment (Liu et al., 2007Liu ZZ, Lozupone C, Hamady M, Bushman FD and Knight R (2007) Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res 35:e120., 2008Liu ZZ, DeSantis TZ, Andersen GL and Knight R (2008) Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res 36:e120.; Hamady et al., 2010Hamady M, Lozupone C and Knight R (2010) Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4:17–27.; Kirchman et al., 2010Kirchman DL, Cottrell MT and Lovejoy C (2010) The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ Microbiol 12:1132–1143.). Therefore, the goal of the present study was to take advantage of pyrosequencing to investigate the identities of members of microbial communities in leachates originating from the decomposition of pig carcasses over a period of 98 days.
Materials and Methods
Testing device
A lab-scale system was designed based on animal carcass mass burial procedures developed by the Korean Ministry for Food, Agriculture, Forestry and Fisheries. The lab-scale system, (1/7 normal size), was 0.54 m (l) x 1.4 m (w) x 0.85 m (h), approximately 0.67 m3 in volume, and is described in detail by Yang et al. (2012)Yang SH, Hong SH, Cho SB, Lim JS, Bae SE, Ahn HK and Lee EY (2012) Characterization of microbial community in the leachate associated with the decomposition of entombed pigs. J Microbiol Biotechn 22:1330–5.. Three lab-scale systems, each of which can accommodate an approximately 115 kg single pig carcass, were placed in a room at 35 °C. The leachate samples were collected using a peristaltic pump from each of three containers once each week for the first month and then at 6 and 14 weeks. Serial collected sample sets from the triplicate system during decomposition were used for molecular analysis. The pH was measured immediately after collection using a bench-top pH meter (Orion 4 STAR, Thermo Scientific Inc., Beverly, USA), and then the leachate samples were stored at −80 °C.
PCR amplification and pyrosequencing
Total genomic DNA was extracted from leachates using a Fast DNA Spin kit for soil (MP Bio, Texas, USA). Fragments of the V1–V3 regions of the 16S rDNA were amplified using fusion primers from genomic DNA extracted from leachate samples (Hur et al., 2011Hur M, Kim Y, Song HR, Kim JM, Choi YI and Yi H (2011) Effect of genetically modified poplars on soil microbial communities during the phytoremediation of waste mine tailings. Appl Environ Microb 77:7611–7619.). Each PCR reaction was performed in a final volume of 50 μL with 5 μLof 10X Taq buffer, dNTP mixture (Takara, Shiga, Japan), 10 μM of each primer and2Uof Taq polymerase (Ex Taq, Takara) using a C1000 Touch thermal cycler (Bio-Rad, Hercules, CA, USA). Amplification reactions were performed as follows: 94 °C for 5 min, 30 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, and a final extension step at 72 °C for 7 min. After visualizing the PCR product using gel electrophoresis and the Gel Doc system (Bio-Rad), the amplicons were purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA, USA). Purified amplicons from different samples (same concentration of each sample) were pooled and purified using an AMPure bead kit (Agencourt Bioscience, Beverly, MA, USA). Pooled products were amplified on the sequencing beads using emulsion PCR, and the beads were deposited on a 454 Picotiter Plate and sequenced using a GS Junior system (Roche, Brandford, CT, USA) according to the manufacturer’s instructions.
Analysis of pyrosequencing
Pyrosequencing analysis was performed according to published methods (Chun et al., 2010Chun J, Kim KY, Lee JH and Choi Y (2010) The analysis of oral microbial communities of wild-type and toll-like receptor 2-deficient mice using a 454 GS FLX Titanium pyrosequencer. BMC Microbiol 10:e101.; Hur et al., 2011Hur M, Kim Y, Song HR, Kim JM, Choi YI and Yi H (2011) Effect of genetically modified poplars on soil microbial communities during the phytoremediation of waste mine tailings. Appl Environ Microb 77:7611–7619.; Kim et al., 2012aKim BS, Kim JN, Yoon SH, Chun J and Cerniglia CE (2012a) Impact of enrofloxacin on the human intestinal microbiota revealed by comparative molecular analysis. Anaerobe 18:310–20.). The raw data obtained from different samples were sorted by their unique barcodes. We discarded sequences containing two or more ambiguous nucleotides (N), low quality score (average score < 25), or reads shorter than 300 base pairs (bp). After deleting chimeric sequences, the taxonomic assignment of each read was conducted using the EzTaxon-e database, which contains representative phylotypes of cultured and uncultured sequences in the GenBank database. The phylogenetic positions were manually edited (Kim et al., 2012bKim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S and Chun J (2012b) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Micr 62:716–721.). The richness and diversity of samples were calculated according to the Chao1 estimation and Shannon index at the 3% dissimilarity level using the Mothur program (Schloss et al., 2009Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb 75:7537–7541.). To compare different read numbers among samples, random subsampling was conducted using CLcommunity software (Chunlab, Inc., Seoul, Korea). The phylogenetic distances between communities were estimated using Fast UniFrac (Hamady et al., 2010Hamady M, Lozupone C and Knight R (2010) Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4:17–27.) and visualized with principal coordinate analysis (PCoA). The significances of the differences between community structures were calculated using Libshuff analysis (Singleton et al., 2001Singleton DR, Furlong MA, Rathbun SL and Whitman WB (2001) Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl Environ Microb 67:4374–4376.). Pyrosequences obtained in this study were uploaded to the EMBL SRA database under the study accession number ERP002343 (http://www.ebi.ac.uk/ena/data/view/ERP002343).
Results and Discussion
The analyses of bacterial communities in leachates from different stages of the decomposition of pig carcasses were conducted by pyrosequencing of 16S rRNA gene amplicons. A total of 51,230 reads were obtained from six different sampling times, and normalized read numbers in each sample were used to compare estimated values among samples (Table 1). The minimum value of Good’s coverage was 81% (week 6 sample), and the maximum value was 90% (week 2 sample). The highest and lowest numbers of operational taxonomic units (OTUs) was observed at week 6 (1,002 OTUs) and 2 (549), respectively. The diversity of community composition changed during the periods of decomposition. The Shannon index decreased from 1 (4.10) to week 4 (3.22), increased at week 6 (4.58), and decreased again at week 14 (3.06). These results indicate that the bacterial communities in leachates at 1 and 6 weeks consisted of various bacterial species and were complex. The presence of dominant bacterial species decreased the diversity of the bacterial community observed during the early stage.
Comparison of pH to the abundance of bacterial 16S rDNA sequences of leachates from decomposing pig carcasses.
The rarefaction curves of the samples are presented in Figure 1. The highest richness of the bacterial community was detected in the 6-week sample and the lowest at 2 and 14 weeks. The dominant phylum was Firmicutes in all samples, and the abundance of Proteobacteria and Bacteroidetes was increased in the 6-week sample (Figure 2). The shift in the composition of the bacterial community was investigated in more detail at the genus level. The abundance of unclassified bacteria within Tissierella genus increased from weeks 2 to 4 (27% of total reads at week 1, > 46.1% from weeks 2 to 4). Peptostreptococcus increased at 14 weeks (38.7% of total reads). Ruminal Peptostreptococcus plays an important role in the degradation of valine, isoleucine, and leucine (Chen and Russell, 1989Chen GJ and Russell JB (1989) Sodium-dependent transport of branched-chain smino-acids by a monensin-sensitive ruminal Peptostreptococcus. Appl Environ Microb 55:2658–2663.). The relative abundance of Anaerosalibacter was the highest at week 1 (over 11% of total reads); this genus was isolated from oil sludge and is reported to be a reducing species during the decomposition process (Rezgui et al., 2012Rezgui R, Maaroufi A, Fardeau ML, Ben Ali Gam Z, Cayol JL, Ben Hamed S and Labat M (2012) Anaerosalibacter bizertensis gen. nov., sp. nov., a new halotolerant bacterium isolated from sludge. Int J Syst Evol Micr 60:2469–2474.).
Rarefaction curves of six different samples obtained from leachates. Each number indicates the time of sampling.
Double pie charts of bacterial communities. The inner and outer pies represent the composition of phyla and genera, respectively. The numbers above each pie indicates the sampling time. The nomenclature of phylotypes is based on the EzTaxon-e database (Kim et al., 2012bKim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S and Chun J (2012b) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Micr 62:716–721.; http://eztaxon-e.ezbiocloud.net/).
Tissierella and Tepidimicrobium are anaerobic species that reduce protein and amino acids, and these bacteria generally increased during decomposition (Harms et al., 1998Harms C, Schleicher A, Collins MD and Andreesen JR (1998) Tissierella creatinophila sp. nov., a Gram-positive, anaerobic, non-spore-forming, creatinine-fermenting organism. Int J Syst Bacteriol 48:983–993.; Slobodkin et al., 2006Slobodkin AI, Tourova TP, Kostrikina NA, Lysenko AM, German KE, Bonch-Osmolovskaya EA and Birkeland NK (2006) Tepidimicrobium ferriphilum gen nov, sp nov, a novel moderately thermophilic, Fe(III)-reducing bacterium of the order Clostridiales. Int J Syst Evol Micr 56:369–372.). Our present data reveal that there was a gradual reduction of Clostridium spp. and Anaerosalibacter from weeks 2 to 14 while uncultured Tissierella spp. (AB175363_g and EF559164_g) and Tepidimicrobium was increased at week 6. The relative abundance of the various genera increased at week 6, and this produced the highest diversity of bacteria. After six weeks the leachate might have contained a variety of organic compounds that supported the growth of diverse classes of bacteria.
The result of the microbial communities grouped by UniFrac and PCoA is shown in Figure 3. The community compositions in the two and four weeks samples were similar, while the compositions changed after six and 14 weeks, which suggests that different bacteria were active at each stage of decomposition. Furthermore, the first principal coordinate (PC1, variance of 30.0%) separated the microbial communities in accordance with the pattern of pH suggesting that major contributor to affect dynamics of the microbial communities might be pH, although other factors affecting microbial community did not be shown in this study.
Differences between bacterial communities at each time point were analyzed and compared using PCoA based on Fast UniFrac distance. The number above each circle indicates the sampling time.
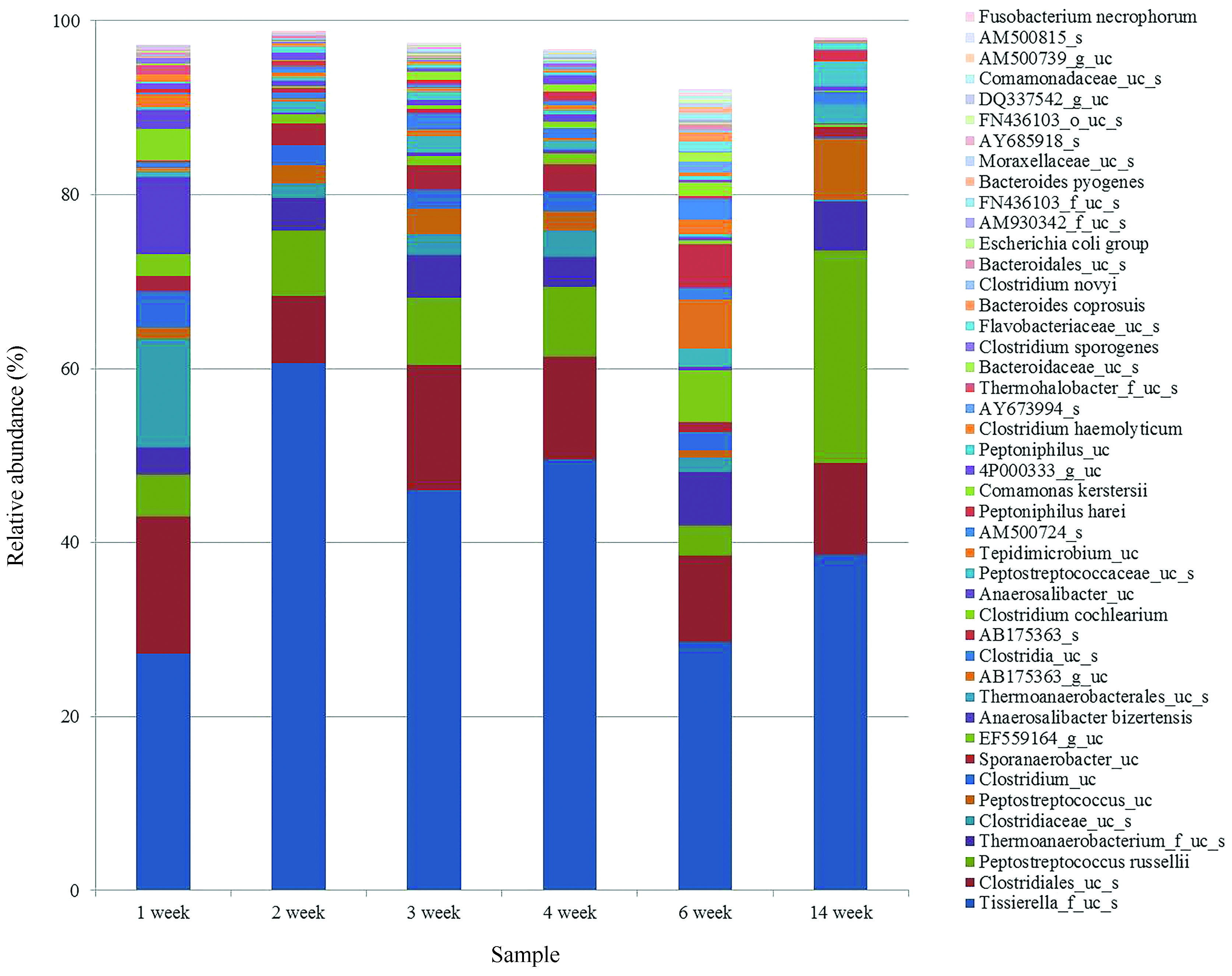
Although most sequences were identified as uncultured bacteria at species, the assignment using the Eztaxon-e database provided detailed information about the uncultured bacteria (Figure. 4). Uncultured bacteria similar to Tissierella (Tissierella_f_uc_s) were the most dominant species among all samples, and most of the sequences identified as uncultured Tissierella were similar to GenBank accession numbers AB175363 and EF559164. Uncultured Tissierella AB175363 is a mesophilic anaerobic protein-degrading bacterium, and uncultured Tissierella EF559164 is involved in the methanization of cellulose under mesophilic conditions (Tang et al., 2005Tang YQ, Shigematsu T, Morimura S and Kida K (2005) Microbial community analysis of mesophilic anaerobic protein degradation process using bovine serum albumin (BSA)-fed continuous cultivation. J Biosci Bioeng 99:150–164.; Li et al., 2009Li TL, Mazeas L, Sghir A, Leblon G and Bouchez T (2009) Insights into networks of functional microbes catalysing methanization of cellulose under mesophilic conditions. Environ Microbiol 11:889–904.). Therefore, uncultured Tissierella detected here may play a main role in the degradation of pig proteins during 14 weeks. The numbers of P. russellii was increased significantly at week 14, and Clostridium spp. were abundant in the week 1 sample. P. russellii was isolated from a swine-manure storage pit, and the presence of this species could be related to the decomposition stage of organic materials (Whitehead et al., 2011Whitehead TR, Cotta MA, Falsen E, Moore E and Lawson PA (2011) Peptostreptococcus russellii sp nov., isolated from a swine-manure storage pit. Int J Syst Evol Micr 61:1875–1879.). Clostridium spp. are ubiquitous anaerobes found in various environments and are potentially pathogenic. Clostridium spp. were isolated from the gastrointestinal tract of pigs, and are abundant at relatively early stages of decomposition (Varel et al., 1995Varel VH, Tanner RS and Woese CR (1995) Clostridium herbivorans sp nov, a cellulolytic anaerobe from the pig intestine. Int J Syst Bacteriol 45:490–494.; Alvarez-Perez et al., 2009Alvarez-Perez S, Alba P, Blanco JL and Garcia ME (2009) Detection of toxigenic Clostridium difficile in pig feces by PCR. Vet Med-Czech 54:360–366.; Garcia et al., 2009Garcia A, Ayuso D, Benitez JM, Garcia WL, Martinez R and Sanchez S (2009) Clostridium novyi infection causing sow mortality in an Iberian pig herd raised in an outdoor rearing system in Spain. J Swine Health Prod 17:264–268.). Although most of the bacteria detected are anaerobic, the proportion of the aerobic bacterium Comamonas kerstersii was increased at week 6. Species of Comamonas are also present during anaerobic acidification of pig excreta (Zhang et al., 2011Zhang DD, Yuan XF, Guo P, Suo YL, Wang XF, Wang WD and Cui ZJ (2011) Microbial population dynamics and changes in main nutrients during the acidification process of pig manures. J Environ Sci (China) 23:497–505.). The diverse uncultured bacteria detected in this study provide further information about leachate decomposition in the environment.
Comparison of the species compositions of bacterial communities during each period. The names of species are based on the EzTaxon-e database.
Recently it was reported that limestone-containing hydrated lime and quicklime affected the decomposition of pig carcasses by changing the pH in burial pits (Schotsmans et al., 2012Schotsmans EM, Denton J, Dekeirsschieter J, Ivaneanu T, Leentjes S, Janaway RC and Wilson AS (2012) Effects of hydrated lime and quicklime on the decay of buried human remains using pig cadavers as human body analogues. Forensic Sci Int 217:50–9.). Quicklime was used in the present study to decompose the pig carcasses, which might have contributed to the sharp increase in pH to 8.79 caused by permeation of the leachate through the layer of quicklime during six weeks (Table 1). This increased pH might be related to the highest level of diversity in this sample. The shifts in bacterial communities indicate that different bacterial species played a role during specific decomposition periods and generated and degraded organic materials that differed during each period.
Investigations of shifts in the population of bacterial communities are important, because the leachate affects the conditions of adjacent land and the surrounding environment. Thus, microorganism can influence the ecosystem balance through the activities of their metabolites (Stahl and Tiedje, 2002Stahl DA and Tiedje JM (2002) Microbial Ecology and Genomics: A Crossroads of Opportunity. Am Acad Microbiol, Washington. DC, 28 pp.). Furthermore, the decomposition of carcasses adversely affects the environment for as long as two to ten weeks after burial (McDaniel, 1991McDaniel HA (1991) Environmental protection during animal disease eradication programmes. Rev Sci Tech OIE 10:867–884.). Our present findings indicate a continual shift in the composition of bacterial communities in leachates generated during the decomposition of pig carcasses. The bacterial community in the leachates could affect the surrounding environment, including soil and groundwater at or near the burial site, potentially leading to changes in the composition of organic materials, indigenous microbes, and symbionts. These changes can be expected to adversely affect public health.
In conclusion, we found shifts in bacterial communities in leachates of decomposing pig carcasses during 14 weeks. Different bacterial groups dominated and may play major roles in the decomposition during specific time intervals. Uncultured Tissiellera spp. and Peptostreptococcus spp. were present in abundance, and the pH was an interactive factor in the decomposition of pig carcasses. The results illustrate the population dynamics of bacterial communities and revealed the predominant bacterial species in such leachates. This information holds promise to improve and implement measures against any future outbreaks of FMD in Korea and other countries and could also be helpful for maintaining the health of ecosystems. Clearly, further studies of burial sites areas will be required to manage and to avert adverse effects on public health.
Acknowledgments
This work was carried out with the support of Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ90711004), Rural Development Administration, Republic of Korea. The authors wish to thank Mrs. Song Ja Jeong for her help on laboratory work.
-
Associate Editor: Carlos F.M. Menck
References
- Ahn HK (2012) 2010 FMD outbreak in Korea: Goverment’s response to this emergency and important lessons learned. Keynote presentation at 4th International Symposium on managing Animal Mortality, Products, By Products, and Associated Health Risk, May 21–24, Dearborn, Michigan, University of Maine.
- Alvarez-Perez S, Alba P, Blanco JL and Garcia ME (2009) Detection of toxigenic Clostridium difficile in pig feces by PCR. Vet Med-Czech 54:360–366.
- Blagodatskaya EV and Anderson TH (1998) Interactive effects of pH and substrate quality on the fungal-to-bacterial ratio and QCO (2) of microbial communities in forest soils. Soil Biol Biochem 30:1269–1274.
- Brooksby JB (1982) Portraits of viruses: foot-and-mouth disease virus. Intervirology 18:1–23.
- Campobasso CP, Di Vella G and Introna F (2001) Factors affecting decomposition and Diptera colonization. Forensic Sci Int 120:18–27.
- Carter DO and Tibbett M (2006) Microbial decomposition of skeletal muscle tissue (Ovis aries) in a sandy loam soil at different temperatures. Soil Biol Biochem 38:1139–1145.
- Carter DO, Yellowlees D and Tibbett M (2007) Cadaver decomposition in terrestrial ecosystems. Naturwissenschaften 94:12–24.
- Chen GJ and Russell JB (1989) Sodium-dependent transport of branched-chain smino-acids by a monensin-sensitive ruminal Peptostreptococcus. Appl Environ Microb 55:2658–2663.
- Christensen TH, Kjeldsen P, Albrechtsen HJ, Heron G, Nielsen PH, Bjerg PL and Holm PE (1994) Attenuation of landfill leachate pollutants in aquifers. Crit Rev Env Sci Tec 24:119–202.
- Chun J, Kim KY, Lee JH and Choi Y (2010) The analysis of oral microbial communities of wild-type and toll-like receptor 2-deficient mice using a 454 GS FLX Titanium pyrosequencer. BMC Microbiol 10:e101.
- Conant RT, Dalla-Betta P, Klopatek CC and Klopatek JA (2004) Controls on soil respiration in semiarid soils. Soil Biol Biochem 36:945–951.
- Domingo E, Mateu MG, Martínez MA, Dopazo J, Moya A and Sobrino F (1990) Genetic variability and antigenic diversity of foot-and-mouth disease virus. In: Kurstak E, Marusyk RG, Murphy SA and VanRegenmortel MHV (eds) Applied Virology Research, vol 2: Virus Variation and Epidemiology. Plenum Publishing Corp, New York, pp 233–266.
- Garcia A, Ayuso D, Benitez JM, Garcia WL, Martinez R and Sanchez S (2009) Clostridium novyi infection causing sow mortality in an Iberian pig herd raised in an outdoor rearing system in Spain. J Swine Health Prod 17:264–268.
- Gill-King H (1997) Chemical and ultrastructural aspects of decomposition. In: Haglund WD and Sorg MH (eds) Forensic Taphonomy: The Postmortem Fate of Human Remains. CRC Press, New York, pp 93–104.
- Gruman MJ and Baxt B (2004) Foot-and-mouth disease. Clin Microbiol Rev 17:465–493.
- Hamady M, Lozupone C and Knight R (2010) Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4:17–27.
- Harms C, Schleicher A, Collins MD and Andreesen JR (1998) Tissierella creatinophila sp. nov., a Gram-positive, anaerobic, non-spore-forming, creatinine-fermenting organism. Int J Syst Bacteriol 48:983–993.
- Hur M, Kim Y, Song HR, Kim JM, Choi YI and Yi H (2011) Effect of genetically modified poplars on soil microbial communities during the phytoremediation of waste mine tailings. Appl Environ Microb 77:7611–7619.
- Keener HM, Elwell DL and Monnin MJ (2000) Procedures and equations for sizing of structures and windrows for composting animal mortalities. Appl Eng Agric 16:681–692.
- Kim BS, Kim JN, Yoon SH, Chun J and Cerniglia CE (2012a) Impact of enrofloxacin on the human intestinal microbiota revealed by comparative molecular analysis. Anaerobe 18:310–20.
- Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S and Chun J (2012b) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Micr 62:716–721.
- Kirchman DL, Cottrell MT and Lovejoy C (2010) The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ Microbiol 12:1132–1143.
- Li TL, Mazeas L, Sghir A, Leblon G and Bouchez T (2009) Insights into networks of functional microbes catalysing methanization of cellulose under mesophilic conditions. Environ Microbiol 11:889–904.
- Liu ZZ, Lozupone C, Hamady M, Bushman FD and Knight R (2007) Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res 35:e120.
- Liu ZZ, DeSantis TZ, Andersen GL and Knight R (2008) Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res 36:e120.
- Ludvigsen L, Albrechtsen HJ, Holst H and Christensen TH (1997) Correlating phospholipid fatty acids (PLFA) in a landfill leachate polluted aquifer with biogeochemical factors by multivariate statistical methods. FEMS Microbiol Rev 20:447–460.
- Lynch JM and Hobbie JE (1988) Micro-Organisms in Action: Concepts and Applications in Microbial Ecology. Blackwell Scientific Publications, Oxford, 363 pp.
- Mann RW, Bass WM and Meadows L (1990) Time since death and decomposition of the human body: variables and observations in case and experimental field studies. J Forensic Sci 35:103–111.
- Matthies C, Erhard HP and Drake HL (1997) Effects of pH on the comparative culturability of fungi and bacteria from acidic and less acidic forest soils. J Basic Microb 37:335–343.
- McDaniel HA (1991) Environmental protection during animal disease eradication programmes. Rev Sci Tech OIE 10:867–884.
- NRC (2002) Biosolids Applied to Land: Advancing Standards and Practices. National Research Council, Washington DC, 344 pp.
- Paul EA and Clark FE (1996) Soil Microbiology and Biochemistry. 2nd edition. Academic Press, San Diego, 340 pp.
- Pfeiffer S, Milne S and Stevenson RM (1998) The natural decomposition of adipocere. J Forensic Sci 43:368–370.
- Pratt DL, Dumonceaux TJ and Fonstad TA (2010) Determination of Microbial Communities beneath Livestock Burial Sites. DOI: 10.13031/2013.36276 Conference: ASABE/CSBE North Central Intersectional Meeting.Saskatoon, SK.
» https://doi.org/10.13031/201336276 - Rezgui R, Maaroufi A, Fardeau ML, Ben Ali Gam Z, Cayol JL, Ben Hamed S and Labat M (2012) Anaerosalibacter bizertensis gen. nov., sp. nov., a new halotolerant bacterium isolated from sludge. Int J Syst Evol Micr 60:2469–2474.
- Röling WFM, van Breukelen BM, Braster M, Lin B and van Verseveld HW (2001) Relationships between microbial community structure and hydrochemistry in a landfill leachate-polluted aquifer. Appl Environ Microb 67:4619–4629.
- Sasaki H, Nonaka J, Otawa K, Kitazume O, Asano R, Sasaki T and Nakai Y (2009) Analysis of the structure of the bacterial community in the livestock manure-based composting process. Asian-Aust J Anim 22:113–118.
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb 75:7537–7541.
- Schotsmans EM, Denton J, Dekeirsschieter J, Ivaneanu T, Leentjes S, Janaway RC and Wilson AS (2012) Effects of hydrated lime and quicklime on the decay of buried human remains using pig cadavers as human body analogues. Forensic Sci Int 217:50–9.
- Seellers RF (1995) Growth and titration of the viruses of foot-and mouth disease and vesicular stomatitis in kidney monolayer tissue cultures. Nature 176:121–124.
- Sharma PK and Das SK (1984) Occurrence of foot-and mouth disease and distribution of virus type in the hill states of North Eastern region of India. Indian J Anim Sci 4:117–118.
- Singleton DR, Furlong MA, Rathbun SL and Whitman WB (2001) Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl Environ Microb 67:4374–4376.
- Slobodkin AI, Tourova TP, Kostrikina NA, Lysenko AM, German KE, Bonch-Osmolovskaya EA and Birkeland NK (2006) Tepidimicrobium ferriphilum gen nov, sp nov, a novel moderately thermophilic, Fe(III)-reducing bacterium of the order Clostridiales. Int J Syst Evol Micr 56:369–372.
- Stahl DA and Tiedje JM (2002) Microbial Ecology and Genomics: A Crossroads of Opportunity. Am Acad Microbiol, Washington. DC, 28 pp.
- Tang YQ, Shigematsu T, Morimura S and Kida K (2005) Microbial community analysis of mesophilic anaerobic protein degradation process using bovine serum albumin (BSA)-fed continuous cultivation. J Biosci Bioeng 99:150–164.
- Tian Y, Yang H, Wu X and Li DT (2005) Molecular analysis of microbial community in a groundwater sample polluted by landfill leachate and seawater. J Zhejiang Univ Sci 6:165–170.
- Van’tHoff JH (1898) Lectures on Theoretical and Physical Chemistry. Part 1. Chemical Dynamics. Edward Arnold, London, pp. 224–229.
- Varel VH, Tanner RS and Woese CR (1995) Clostridium herbivorans sp nov, a cellulolytic anaerobe from the pig intestine. Int J Syst Bacteriol 45:490–494.
- Vass AA, Bass WM, Wolt JD, Foss JE and Ammons JT (1992) Time since death determinations of human cadavers using soil solution. J Forensic Sci 37:1236–1253.
- Whitehead TR, Cotta MA, Falsen E, Moore E and Lawson PA (2011) Peptostreptococcus russellii sp nov., isolated from a swine-manure storage pit. Int J Syst Evol Micr 61:1875–1879.
- Woodbury EL (1995) A review of the possible mechanisms for the persistence of foot-and-mouth disease virus. Epidemiol Infect 114:1–13.
- Yang SH, Hong SH, Cho SB, Lim JS, Bae SE, Ahn HK and Lee EY (2012) Characterization of microbial community in the leachate associated with the decomposition of entombed pigs. J Microbiol Biotechn 22:1330–5.
- Zhang DD, Yuan XF, Guo P, Suo YL, Wang XF, Wang WD and Cui ZJ (2011) Microbial population dynamics and changes in main nutrients during the acidification process of pig manures. J Environ Sci (China) 23:497–505.
Internet Resources
- EzTaxon-e database, http://eztaxon-e.ezbiocloud.net (accessed at 2012.10.02).
» http://eztaxon-e.ezbiocloud.net
Publication Dates
-
Publication in this collection
Sept 2015
History
-
Received
14 Sept 2014 -
Accepted
10 Feb 2015