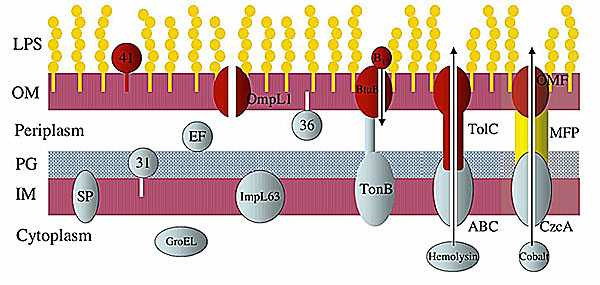
We report novel features of the genome sequence of Leptospira interrogans serovar Copenhageni, a highly invasive spirochete. Leptospira species colonize a significant proportion of rodent populations worldwide and produce life-threatening infections in mammals. Genomic sequence analysis reveals the presence of a competent transport system with 13 families of genes encoding for major transporters including a three-member component efflux system compatible with the long-term survival of this organism. The leptospiral genome contains a broad array of genes encoding regulatory system, signal transduction and methyl-accepting chemotaxis proteins, reflecting the organism's ability to respond to diverse environmental stimuli. The identification of a complete set of genes encoding the enzymes for the cobalamin biosynthetic pathway and the novel coding genes related to lipopolysaccharide biosynthesis should bring new light to the study of Leptospira physiology. Genes related to toxins, lipoproteins and several surface-exposed proteins may facilitate a better understanding of the Leptospira pathogenesis and may serve as potential candidates for vaccine.
Leptospira interrogans; Outer membrane proteins; Lipopolysaccharides; Transport; Regulatory systems