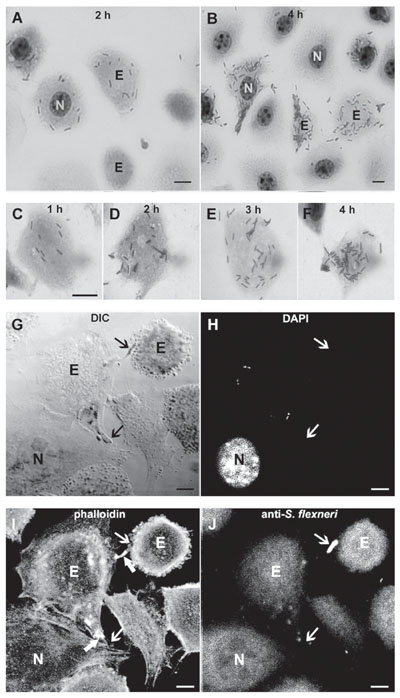
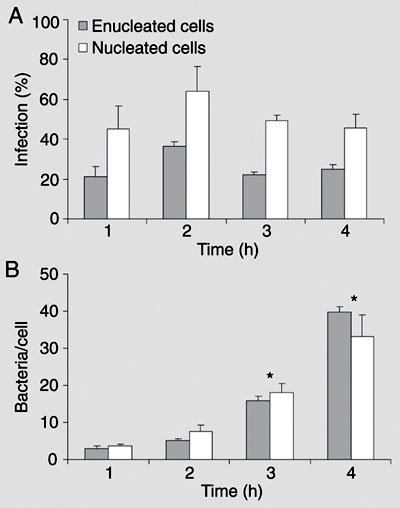
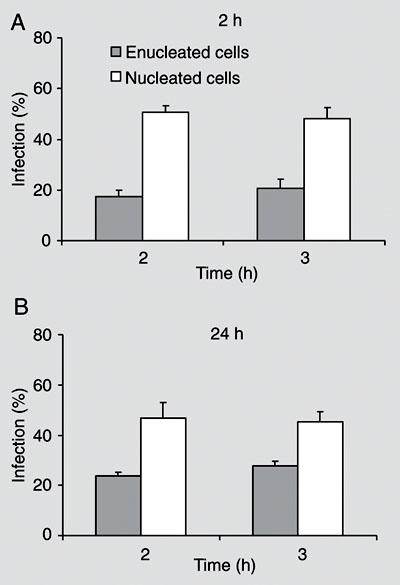
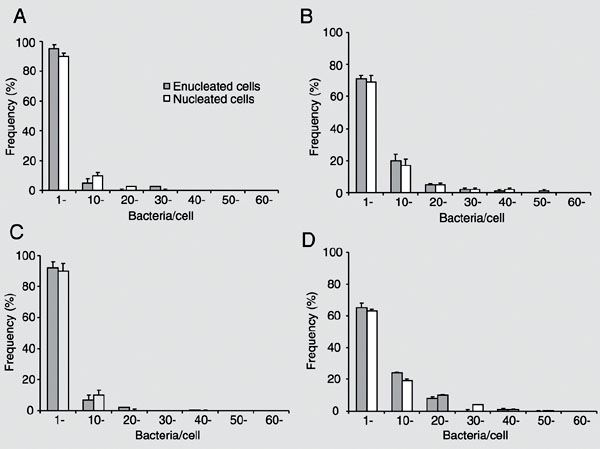
Invasive bacteria can induce their own uptake and specify their intracellular localization; hence it is commonly assumed that proximate modulation of host cell transcription is not required for infection. However, bacteria can also modulate, directly or indirectly, the transcription of many host cell genes, whose role in the infection may be difficult to determine by global gene expression. Is the host cell nucleus proximately required for intracellular infection and, if so, for which pathogens and at what stages of infection? Enucleated cells were previously infected with Toxoplasma gondii, Chlamydia psittaci, C. trachomatis, or Rickettsia prowazekii. We enucleated L929 mouse fibroblasts by centrifugation in the presence of cytochalasin B, and compared the infection with Shigella flexneri M90T 5a of nucleated and enucleated cells. Percent infection and bacterial loads were estimated with a gentamicin suppression assay in cultures fixed and stained at different times after infection. Enucleation reduced by about half the percent of infected cells, a finding that may reflect the reduced endocytic ability of L929 cytoplasts. However, average numbers of bacteria and frequency distributions of bacterial numbers per cell at different times were similar in enucleated and nucleated cells. Bacteria with actin-rich tails were detected in both cytoplasts and nucleated cells. Lastly, cytoplasts were similarly infected 2 and 24 h after enucleation, suggesting that short-lived mRNAs were not involved in the infection. Productive S. flexneri infection could thus take place in cells unable to modulate gene transcription, RNA processing, or nucleus-dependent signaling cascades.
Shigella flexneri; Cell enucleation; Cytoplasts; L929 fibroblasts