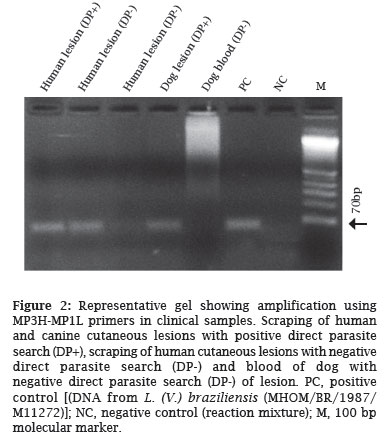
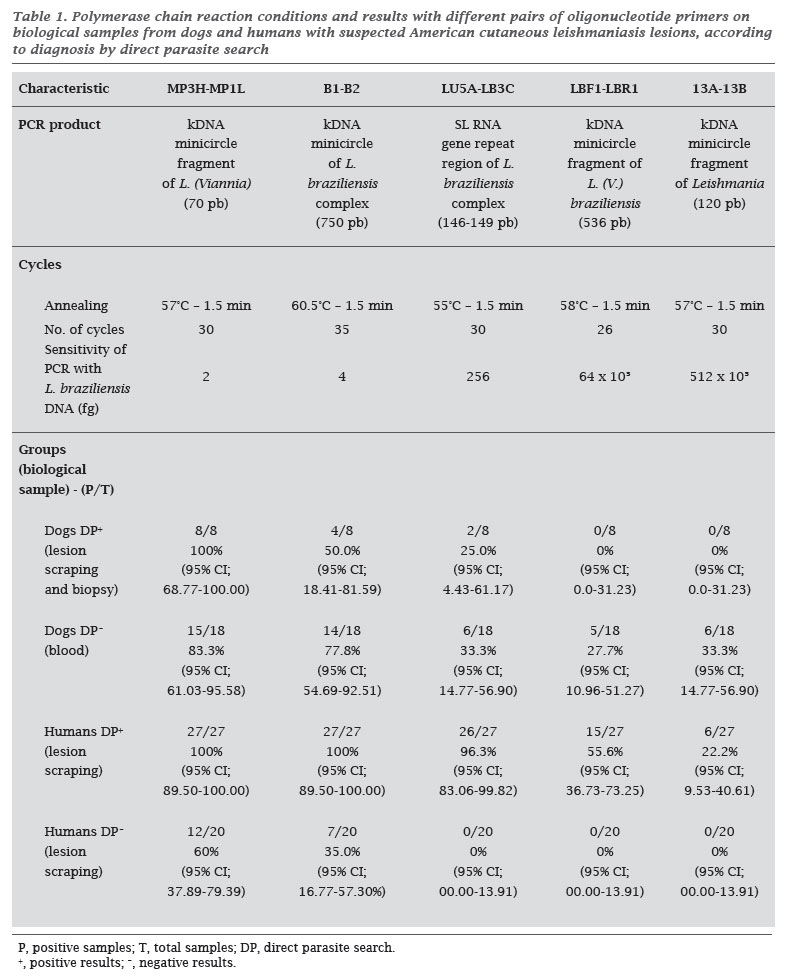
OBJECTIVE: The objective of this study was to analyze different primers that are commonly used in epidemiological studies for the detection of Leishmania DNA by PCR, and to compare them to the conventional direct parasite search for American cutaneous leishmaniasis (ACL) diagnosis. MATERIAL AND METHODS: Five pairs of primers, four of them derived from Leishmania kDNA sequences (MP3H-MP1L; B1-B2; LBF1-LBR1; 13A-13B), and one derived from the SL RNA (mini-exon) gene repeat (LU5A-LB3C), reported previously, were used. RESULTS: The MP3H-MP1L primers were the best at amplifying the DNA, detecting 2 fg of Leishmania spp. DNA. The 13A-13B primers presented the worst performance, detecting 512 x 10³ fg of DNA. CONCLUSION: The wide variation in the analytical sensitivity of the primers used in the PCR, and the significant differences from the conventional method of ACL diagnosis found in this study, emphasize the importance of standardizing the PCR technique, analyzing sensitivity, and selecting suitable oligonucleotide primers.
leishmaniasis; polymerase chain reaction; Leishmania; DNA primers