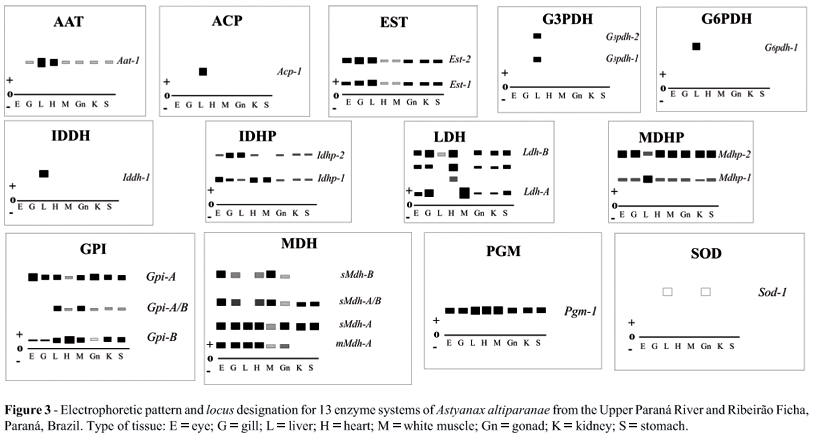
Allozyme data was used to assess the genetic diversity Astyanax altiparanae populations from the floodplain of the Upper Paraná River (PR). Specimens were collected in the southern Brazilian state of Paraná from PR in Porto Rico municipality and Ribeirão Ficha (RF) in Ubiratã municipality. The authors used 15% (w/v) corn starch gel electrophoresis to identify 21 putative loci for 13 enzymatic systems: Aspartate aminotransferase, 2.6.1.1 (AAT), Acid phosphatase, 3.1.3.2 (ACP), Esterase, 3.1.1.1 (EST), Glycerol-3-phosphate dehydrogenase, 1.1.1.8 (G3PDH), Glucose-6-phosphate dehydrogenase, 1.1.1.49 (G6PDH), Glucose-6-phosphate isomerase, 5.3.1.9 (GPI), Iditol dehydrogenase, 1.1.1.14 (IDDH), Isocitrate dehydrogenase - NADP+, 1.1.1.42 (IDH), L-Lactate dehydrogenase, 1.1.1.27 (LDH), Malate dehydrogenase, 1.1.1.37 (MDH), Malate dehydrogenase - NADP+, 1.1.1.40 (MDHP), Phosphoglucomutase, 5.4.2.2 (PGM), and Superoxide dismutase, 1.15.1.1 (SOD). The proportion of polymorphic loci were estimated as 52.38% in the PR population and 38.10% in the RF population. Expected estimated heterozygosities were 0.1518 ± 0.0493 for the PR population and 0.0905 ± 0.0464 for the RF population. The A. altiparanae heterozygosity data were similar to previous estimates for other PR basin characid species. Allele frequencies were significantly different between the PR and RF populations in respect to some loci (Acp-1, G3pdh-1, Gpi-A, Iddh-1, Mdhp-1 and Mdhp-2). Wright’s statistics for all loci were estimated as Fis = 0.3919, Fit = 0.4804 and Fst = 0.1455. Our results show that the A. altiparanae populations studied are genetically different and have a high degree of genetic variability.
allozymes; Astyanax; fishes; genetic variability; Paraná River floodplain