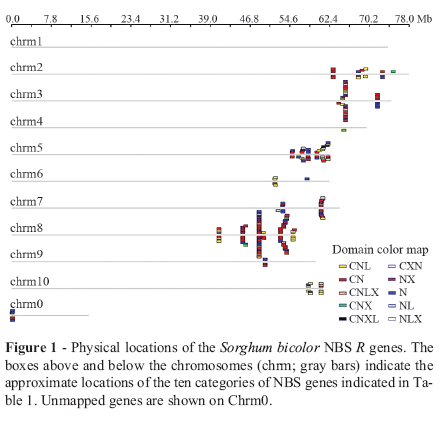
A large set of candidate nucleotide-binding site (NBS)-encoding genes related to disease resistance was identified in the sorghum (Sorghum bicolor) genome. These resistance (R) genes were characterized based on their structural diversity, physical chromosomal location and phylogenetic relationships. Based on their N-terminal motifs and leucine-rich repeats (LRR), 50 non-regular NBS genes and 224 regular NBS genes were identified in 274 candidate NBS genes. The regular NBS genes were classified into ten types: CNL, CN, CNLX, CNX, CNXL, CXN, NX, N, NL and NLX. The vast majority (97%) of NBS genes occurred in gene clusters, indicating extensive gene duplication in the evolution of S. bicolor NBS genes. Analysis of the S. bicolor NBS phylogenetic tree revealed two major clades. Most NBS genes were located at the distal tip of the long arms of the ten sorghum chromosomes, a pattern significantly different from rice and Arabidopsis, the NBS genes of which have a random chromosomal distribution.
bioinformatics; disease resistance gene; nucleotide binding site; Sorghum bicolor