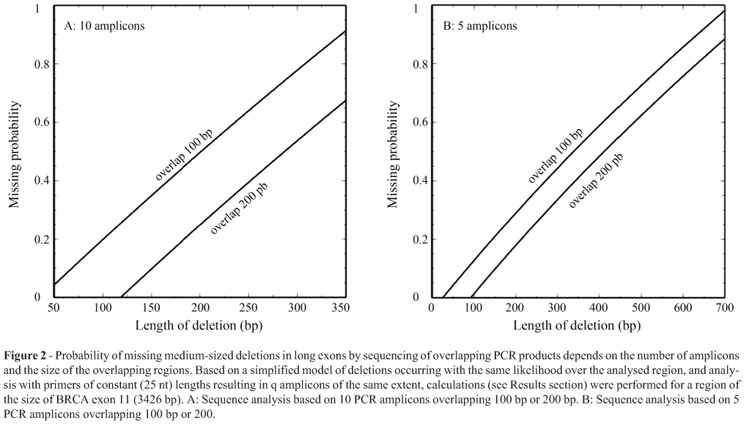
We describe a family with a history of breast and ovarian cancer in which MLPA analysis of the BRCA1 gene pointed to a deletion including a part of exon 11. Further characterization confirmed a loss of 374 bp in a region completely covered by conventional sequencing which had not revealed the deletion. Because this alteration was only detected serendipitously with an MLPA probe, we calculated the probabilities of detecting medium-sized deletions in large exons by methods including initial PCR amplification. This showed that a considerable fraction of medium-sized deletions are undetectable by currently used standard methods of mutation analyses. We conclude that long, widely overlapping amplicons should be used to minimize the risk of missing medium-sized deletions. Alternatively, large exons could be completely covered by narrow-spaced MLPA probes.
mutation analysis; DNA sequencing; PCR; hereditary breast cancer; model calculations