Abstract
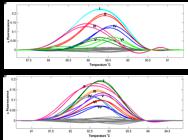
The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) technology has recently emerged as a powerful genomic editing tool with great potential for crop breeding. However, commonly used protocols for screening of CRISPR/Cas9-induced mutations are laborious, time-consuming, and costly. In the present study we examined the applicability of high resolution melting (HRM) analysis fast screening CRISPR/Cas9-induced mutations in T0 plants and subsequent genotyping of T1 populations. Comparative analysis demonstrated that HRM analysis could identify mutant T0 plants carrying various types of mutation, including single nucleotide substitutions and short insertion/deletions, with false positive/negative rates of 0-2.78%. Furthermore, T1 plants derived from single T0 plants could be correctly genotyped by HRM analysis using WT parents and T0 plants as controls. We hence recommend the adoption of HRM analysis in CRISPR/Cas9 mediated genetic studies and breeding in rice and other crop species.
Key words:
Genomic editing; mutation screening; OsLCT1; bell

 Thumbnail
Thumbnail
 Thumbnail
Thumbnail

