Abstracts
The objective of this work was to characterize the grape germplasm in Santa Catarina, Brazil, using microsatellite DNA markers (simple sequence repeats - SSR). The DNA samples were collected from leaves and shoots of accessions of public and private collections from the counties Urussanga, Nova Trento, Rodeio, São Joaquim, Campos Novos, Videira, and Água Doce. Ten SSR loci (VVS2, VVMD5, VVMD7, VVMD27, VrZAG62, VrZAG79, VVMD25, VVMD28, VVMD31, and VVMD32) were analysed by capillary electrophoresis. Molecular profiling was conducted for 190 grapevines (European, American, and hybrids), and 67 genotypes were obtained. The data were compared with each other and with those from the literature and from online databases, in order to identify varieties and discover cases of synonymy and homonymy. Forty molecular profiles corresponded to known varieties, while 27 genotypes were described for the first time. The existence of typical germplasm composed mainly of American and hybrid varieties is an important finding for local viticulture. Applications of the results rely on quality control and certification at the nursery level. Increasing precision in the characterization of grapevine genotypes may help breeding programs.
Vitis spp.; cultivar identification; genetic variability; germplasm collection; molecular markers
O objetivo deste trabalho foi caracterizar a diversidade de videiras em Santa Catarina, por meio de marcadores moleculares microssatélites ("simple sequence repeats" - SSR). Amostras de DNA foram coletadas a partir de folhas e ramos de acessos de coleções de germoplasma públicas e privadas, nos municípios de Urussanga, Nova Trento, Rodeio, São Joaquim, Campos Novos, Videira e Água Doce. Dez loci SSR (VVS2, VVMD5, VVMD7, VVMD27, VrZAG62, VrZAG79, VVMD25, VVMD28, VVMD31 e VVMD32) foram analisados por eletroforese capilar. Foram produzidos perfis moleculares de 190 acessos de videira (europeus, americanos e híbridos), e 67 genótipos foram individualizados. Os dados foram comparados entre si e com aqueles disponíveis em literatura e em bancos de dados online, para a identificação de correspondências e casos de sinonímia e homonímia. Quarenta perfis moleculares corresponderam a variedades conhecidas, e 27 genótipos foram descritos pela primeira vez. A existência de um germoplasma típico, composto principalmente de variedades americanas e híbridas, é um fator importante para a viticultura local. A aplicação desses resultados poderá contribuir para o controle de qualidade e a certificação de mudas. Além disso, aumentar a precisão no que tange à caracterização genética da videira, auxiliará os programas de melhoramento genético.
Vitis spp.; identificação de cultivar; variabilidade genética; coleção de germoplasma; marcadores moleculares
GENETICS
Molecular characterization of grapevine from Santa Catarina, Brazil, using microsatellite markers
Caracterização molecular de videiras de Santa Catarina por marcadores microssatélites
Mariane Ruzza SchuckI; Flavia Maia MoreiraII; Miguel Pedro GuerraI; José Afonso VoltoliniI; Maria Stella GrandoII; Aparecido Lima da SilvaI
IUniversidade Federal de Santa Catarina, Centro de Ciências Agrárias, Programa de Pós-Graduação em Recursos Genéticos Vegetais, Departamento de Fitotecnia, Caixa Postal 476, CEP 88040-900 Florianópolis, SC, Brazil. E-mail: schuck337@gmail.com, mpguerra@cca.ufsc.br, voltolini@cca.ufsc.br, alsilva@cca.ufsc.br
IIFondazione Edmund Mach, Istituto Agrario di San Michele all'Adige, Via E. Mach 1, 38010 San Michele a/A, TN, Italy. E-mail: flavia.moreira@iasma.it, stella.grando@iasma.it
ABSTRACT
The objective of this work was to characterize the grape germplasm in Santa Catarina, Brazil, using microsatellite DNA markers (simple sequence repeats - SSR). The DNA samples were collected from leaves and shoots of accessions of public and private collections from the counties Urussanga, Nova Trento, Rodeio, São Joaquim, Campos Novos, Videira, and Água Doce. Ten SSR loci (VVS2, VVMD5, VVMD7, VVMD27, VrZAG62, VrZAG79, VVMD25, VVMD28, VVMD31, and VVMD32) were analysed by capillary electrophoresis. Molecular profiling was conducted for 190 grapevines (European, American, and hybrids), and 67 genotypes were obtained. The data were compared with each other and with those from the literature and from online databases, in order to identify varieties and discover cases of synonymy and homonymy. Forty molecular profiles corresponded to known varieties, while 27 genotypes were described for the first time. The existence of typical germplasm composed mainly of American and hybrid varieties is an important finding for local viticulture. Applications of the results rely on quality control and certification at the nursery level. Increasing precision in the characterization of grapevine genotypes may help breeding programs.
Index terms:Vitis spp., cultivar identification, genetic variability, germplasm collection, molecular markers.
RESUMO
O objetivo deste trabalho foi caracterizar a diversidade de videiras em Santa Catarina, por meio de marcadores moleculares microssatélites ("simple sequence repeats" - SSR). Amostras de DNA foram coletadas a partir de folhas e ramos de acessos de coleções de germoplasma públicas e privadas, nos municípios de Urussanga, Nova Trento, Rodeio, São Joaquim, Campos Novos, Videira e Água Doce. Dez loci SSR (VVS2, VVMD5, VVMD7, VVMD27, VrZAG62, VrZAG79, VVMD25, VVMD28, VVMD31 e VVMD32) foram analisados por eletroforese capilar. Foram produzidos perfis moleculares de 190 acessos de videira (europeus, americanos e híbridos), e 67 genótipos foram individualizados. Os dados foram comparados entre si e com aqueles disponíveis em literatura e em bancos de dados online, para a identificação de correspondências e casos de sinonímia e homonímia. Quarenta perfis moleculares corresponderam a variedades conhecidas, e 27 genótipos foram descritos pela primeira vez. A existência de um germoplasma típico, composto principalmente de variedades americanas e híbridas, é um fator importante para a viticultura local. A aplicação desses resultados poderá contribuir para o controle de qualidade e a certificação de mudas. Além disso, aumentar a precisão no que tange à caracterização genética da videira, auxiliará os programas de melhoramento genético.
Termos para indexação:Vitis spp., identificação de cultivar, variabilidade genética, coleção de germoplasma, marcadores moleculares.
Introduction
The Brazilian viticulture occupies 89.9 thousand hectares and yields about 1.3 million tons of grapes per year, concentrated in the south, southwest and northeast regions. The state of Rio Grande do Sul produces 90% of vines, juices and other grape products (IBGE, 2007).
The state of Santa Catarina produces the second largest crop, with approximately 50,000 tons of grapes per year. Wines are produced mostly from Vitis labrusca, an American species, or from hybrids adapted to the environmental conditions of the state. Pinheiro Preto, Videira, Tangará, Iomerê, Caçador, Urussanga, Rodeio, and Nova Trento are the main grape-growing counties in Santa Catarina.
Since 1998, viticulture in Santa Catarina has undergone intense transformation, with the establishment of new vineyards of European grapes (V. vinifera). New viticultural areas have been established in the highland regions above 900 m, where climatic conditions determine specific features for grape maturation and wine specificity and quality. The most promising new areas are located in São Joaquim, Bom Retiro, Campos Novos, and Água Doce counties (Schuck et al., 2008).
Unlike the European market, where the "terroir" determines the quality of wines, the Brazilian market is based on the consumption of wines from a single variety. These are classified as regular wines, obtained from American andhybridgrapes,and fine table wines obtained from European white and black grape varieties, mainly Chardonnay, Cabernet Sauvignon, Cabernet Franc, Pinot Noir, and Merlot. The correct identification of grapevine germplasm is important since the expected quality of wines is related to characteristics of the cultivar.
In order to distinguish grape varieties, DNA-based markers offers an advantage over morphological descriptors, as they are less prone to being affected by the environment or the developmental stage (Sefc et al., 2001).
The available markers differ in the methodologies used to detect variability at the DNA level. Since 1990, simple sequence repeats (SSR) markers have been increasingly used as molecular descriptors in grape. Their usefulness has been widely demonstrated in the identification and characterization of stock and rootstock varieties, evaluation of genetic variability, pedigree studies, and genetic mapping (Bowers et al., 1999a; Pollefeys & Bousquet, 2003; Adam-Blondon et al., 2004; Riaz et al., 2004; Costantini et al., 2005). The analysis of grape plants with the aid of SSR markers allows identification profiles to be established and varietal reference data banks to be developed (Dettweiller et al., 1998; Lefort & Roubelakis-Angelakis, 2001; Grando et al., 2002). Access to data banks allows information to be shared and contributes to international cooperation to correctly identify grape germplasm.
The aim of this work was to characterize the diversity of the grape germplasm held in public and private collections in Santa Catarina, Southern Brazil, by means of SSR markers.
Materials and Methods
Leaves and shoots of 183 accessions were collected from 86 European, American and hybrid varieties, and another 7 accessions from unidentified sources (Table 1). The samples were taken from germplasm collections of different regions of Santa Catarina: families Quarezemin, Fellipi, Trevisol, Possamai, and Damiani (private collections) and Epagri (public collection) from Urussanga (28º31'4"S, 49º19'15"W, at altitude 49 m) and from Nova Trento (27º16'60"S, 48º55'0"W, at altitude 30 m); Vinicola San Michele from Rodeio (26º51'5"S, 49º19'60"W, at altitude 106 m); Terras Altas, Suzin, Villa Francioni, and Quinta da Neve (private collections) from São Joaquim (28º18'5"S, 49º55'60"W, at altitude 1,353 m); Epagri (public collection) from Campos Novos (27º23'60"S, 51º12'0"W, at altitude 947 m); Vinicola Pancieri (private collection), and Epagri (public collection) from Videira (27º0'5"S, 51º7'60"W, at altitude 750 m); and Fazenda Boa Esperança (private collection) from Água Doce (27º0'0"S, 51º32'60"W, at altitude 969 m).
DNA extraction was done according to Doyle & Doyle (1990), with the following modifications: freeze-dried material (instead of fresh material) was ground and RNase was added to isopropanol during the DNA precipitation phase. Quantification of the DNA extracts was performed in agarose gel (0.8%), and the samples were then diluted to a concentration of 20 ng µL-1, at the Laboratório de Fisiologia do Desenvolvimento Genético Vegetal, of the Departamento de Fitotecnia, Universidade Federal de Santa Catarina.
In order to facilitate the comparison with data from the literature and international databases, the samples were analyzed at the ten microsatellite loci most frequently used by the international scientific community: VVS2 (Thomas & Scott, 1993), VVMD5, VVMD7 (Bowers et al., 1996), VVMD25, VVMD27, VVMD28, VVMD31, VVMD32 (Bowers et al., 1999b), VrZAG62 and VrZAG79 (Sefc et al., 1999).
Polymerase chain reaction (PCR) was carried out in a 12.5-µL total volume containing 25 mmol L-1 of each dNTP, 0.5 mmol L-1 of each primer, 0.5 U Taq DNA polymerase (GoldTaq - Applied Biosystem, Foster City, CA, USA or BioTaq - Bioline, London, UK), 1X buffer solution, 1.5 mmol L-1 MgCl2, and 20 ng of genomic DNA for each sample. The cycling program used was: denaturation of DNA and activation of Taq DNA polymerase at 95ºC for 7 min (GoldTaq) or 3 min (BioTaq); 35 cycles of amplification distributed in 45 s at 94ºC, 45 s at 50ºC (VVS2, VVMD5, VVMD7, VVMD27, VrZAG62, and VrZAG79) or 56ºC (VVMD25, VVMD28, VVMD31, and VVMD32), 1 min and 30 s at 72ºC; final extension of 7 min at 72ºC; cooling at 4ºC.
Capillary electrophoresis was used to determine the size of alleles. Separation of the microsatellite fragments was performed using the ABI 3100 sequencer and GeneScan 3.7 software (Applied Biosystems). The availability of primers labeled with different fluorescent dyes (Ned, Hex or Fam) enabled the product of different PCR reactions to be loaded in multiplexes. Processing of the data generated by the sequencer was done with the Genotyper 3.7 program (Applied Biosystems). All tests were performed using DNA of the cultivar Chardonnay as a reference with respect to both the quality of amplification and the size of the amplified alleles, allowing the data to be compared with those available in the literature and in databases. This phase of the work was carried out at the Laboratory of Molecular Genetics of the Fondazione Edmund Mach - Istituto Agrario di San Michele all'Adige, in San Michele all'Adige, TN, Italy.
The molecular profiles obtained in this study were compared with online databases: Grape Microsatellite Collection (GMC) - http://www.ismaa.it/areabioav/gmc.html; Greek Vitis Database - (GVD) http://www.biology.uch.gr/gvd; Swiss Vitis Microsatellite Database (SVMD) - http:/www.unine.ch/nccr/svmd/; and GENRO - http://www.genres.de/eccdb/vitis/. Results were also compared to data from the literature not available online from the United States, Switzerland, Italy, Turkey, Armenia, and Georgia (Vouillamoz J., unpublished data). The latter databases currently contain details of microsatellites from more than 2,000 grapevine cultivars.
The Identity program (Wagner & Sefc, 1999) was used to calculate the number of alleles (n), the allele frequencies, expected (He) and observed (Ho) heterozygosity, the estimated frequency of null alleles (r), and the probability of identity (PI) at each locus. This software was also used to detect identical genotypes. Genetic similarity between the genotypes was calculated on the basis of the proportion of alleles in common (DPS), using the Microsat software (Minch et al., 1997). A neighbor-joining dendrogram was constructed from a matrix of genetic divergence (D) between the genotypes, calculated as D = 1-DPS, using Phylip (Felsenstein, 1989) and viewed with Tree View (Page, 1996).
Results and Discussion
The 190 grapevine accessions, belonging to 86 assumed varieties, generated 67 different profiles when analyzed at the 10 SSR loci. The reduction in the number of individual molecular profiles is due to synonymy, accessions identified with different names but having the same molecular profile. Seven synonymy groups were elucidated in this study: (1) Moscato Giallo, Moscato EMBRAPA, and Lorena; (2) Bordô, Grano d'Oro, and São João; (3) Goethe Primo, Goethe Clássica, and Moscatel; (4) Isabel, Isabel Precoce, and Bizaraqui-SM; (5) Itália, Itália Export, Itália Koga, Rubi Itália, Benitaka, Brasil, Redmeire, Lade, and Família Moscatel; (6) Niágara Branca, and Niágara Rosada; and (7) Ora E and Prima E.
Cases of synonymy could be clones of the same variety, which show phenotypic differences (Vignani et al., 1996; Walker et al., 2006). Despite these evident differences, they are not detectable by a low number of SSR markers, which are located in noncoding regions of the genome (Zulini et al., 2005). This was the case of 'Grano D'Oro', a mutation of the cultivar Bordô, which resulted in changes in vigor, productivity, and rusticity. This was also the case of 'Goethe Primo', a variant with white berries from 'Classic Goethe', which has pinkish berries.
Comparison of the data showed some cases of homonymy, that is, clusters of varieties having identical or similar names but different genotypes. This was the case of 'Isabel-SM' accession and the group of synonyms mentioned above, consisting of 'Isabel', 'Isabel Precoce', Catawba S, and Catawba T.
In addition, ten cases of misnaming were brought to light. An accession of 'Muscat of Alexandria' corresponded to the cultivar Sangiovese. 'Pinot Noir' collected in São Joaquim was actually 'Cabernet Sauvignon' and a 'Cabernet Sauvignon' accession of Campos Novos was identical to 'Carmènere'. The accession 'Luan Blanc' from São Joaquim and 'Tempranillo' from Água Doce were both 'Sauvignon Blanc'. Two accessions of 'Trincadeira' collected in São Joaquim and Rodeio corresponded to 'Pinot Noir'. The 'Tinta Roris' was identical to 'Tempranillo', and 'Trincadeira' from Água Doce was actually 'Viognier'. A 'Traminer' accession was wrongly named 'Gewürztraminer'.
Comparison between the molecular profiles from the ten SSR markers employed in the present work and those available in data banks allowed the recognition of 40 from the 67 distinct genotypes. They were mainly well-known international wine varieties. This comparison also allowed the identification of additional five cases of synonymy with known international varieties. Moreover, seven accessions without variety name in the collection were identified (Table 2).
The SSR profiles for rootstocks VR 043-43, 101-14 Mgt, Paulsen 1103, and SO4 did not correspond to the molecular characterization of the same rootstocks by Andrés et al. (2007). The possibility of errors in the collecting procedures or in the introduction of these materials to the collections should be considered.
The remaining 27 molecular profiles did not correspond to those found in the literature, nor in the data banks (Table 3). The identification and description of unique molecular profiles is an important aspect of the regional viticulture. Nineteen genotypes were described for the first time in this study. Most of them are interspecific hybrids developed in Brazil. In other cases, are old varieties introduced during the first wave of Italian immigration to Southern Brazil at the end of the 19th century. Old varieties include the rootstock PESC 2 ("porta-enxerto Santa Catarina") from Rodeio, as well as the well-known 'Goethe' cultivar from Urussanga (Schuck et al., 2008). However, unique SSR profiles were also obtained from accessions thought to belong to international varieties as Catawba, Gros Manseng, and Paulsen 1103, which were determined to be not true-to-type.
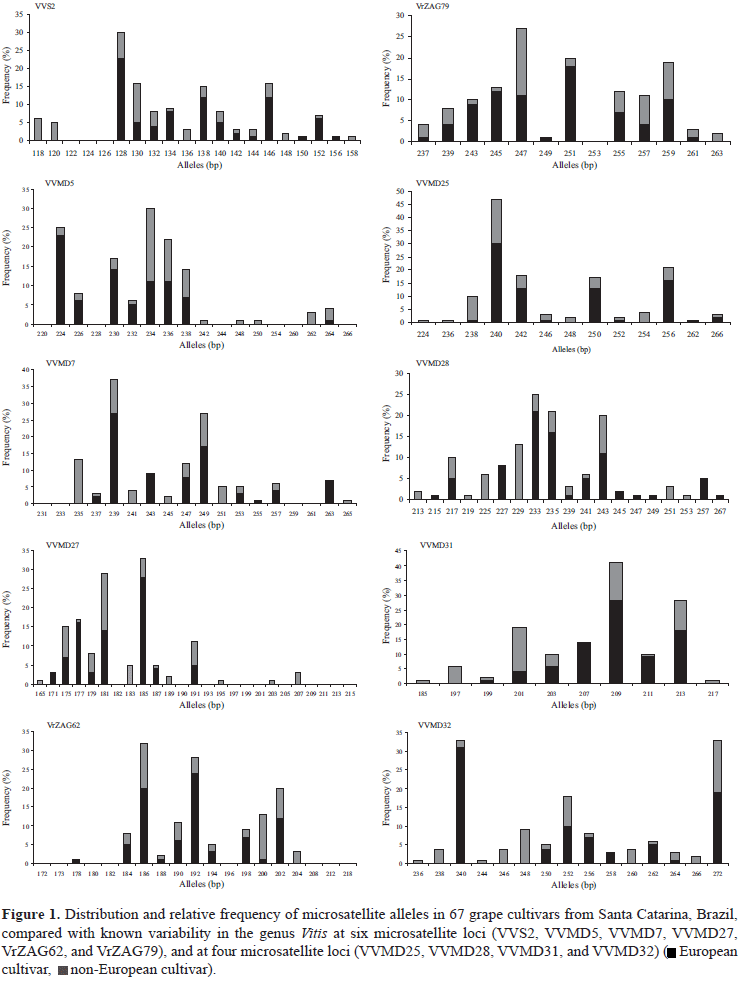
The number of alleles, the expected and observed heterozygosity, the estimated frequency of null alleles, and the probability of identity with the 67 genotypes are listed in Table 4. The germplasm analyzed in this study showed wide variability in the size of SSR alleles and encompassed most (79.9% mean) of the known allelic variants (This et al., 2004). Some alleles commonly present in European varieties were observed, while others had the characteristics of American or hybrid varieties (Figure 1).
The expected heterozygosity, in the 10 evaluated loci, ranged from 0.798 to 0.889, these values being higher than those observed by Sefc et al. (2000) in European varieties (0.677-0.819), but similar to those found in Spanish varieties and Franco-American hybrids (Martín et al., 2003), and in American hybrids cultivated in the USA (Pollefeys & Bousquet, 2003). These results were expected since the grape germplasm from Santa Catarina encompasses European and American varieties, as well as different hybrids.
The observed heterozygosity ranged from 0.687 to 0.849, with values lower than those expected from randomized union of gametes (He) in all loci, except for VVMD7. For these loci, the probability of null alleles was positive, suggesting that most of the apparent homozygotes could be heterozygotes, one allele being visible and the other not. These types of null alleles can occur, when mutations do not allow the linking of primers to the target region (Pollefeys & Bousquet, 2003).
In eight out of ten analyzed loci, the probability of identity was higher than 0.05, and their use in combination resulted in an accumulated probability (the probability of obtaining by chance two identical profiles, when belonging to distinct varieties) of 2.13x10-12, thus confirming the high degree of polymorphism in the chosen SSR markers.
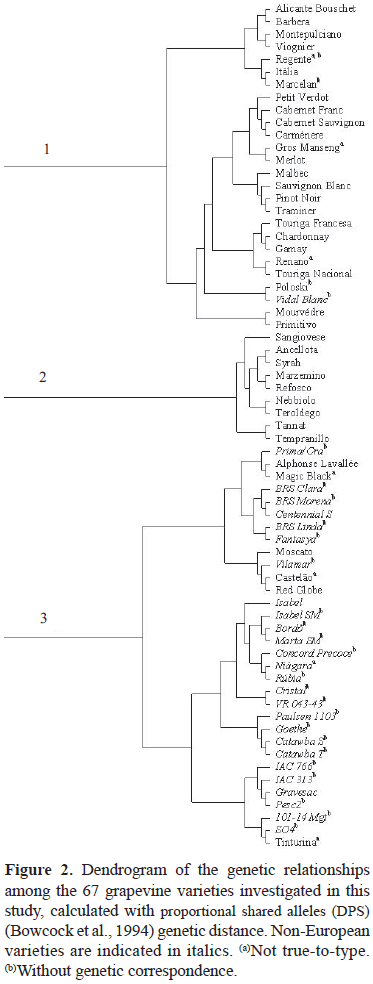
The different molecular profiles were also evaluated to visualize similar groups to check whether such groups coincide with the known history of the varieties. The dendrogram (Figure 2) shows three main differentiated groups, which suggests three different origins for the grape germplasm in Santa Catarina. Group 1 includes European varieties of French origin, such as Cabernet Franc, Cabernet Sauvignon, Carmènere, Merlot, Sauvignon Blanc, Pinot Noir, and Chardonnay. Group 2 is composed by a small number of accessions, mainly Italian varieties, such as Sangiovese, Ancellota, Syrah, Marzemino, Refosco, Nebbiolo, and Teroldego. Finally, the third and largest group comprises almost exclusively American and hybrid varieties, such as Prima/Ora, BRS Clara, BRS Morena, Centennial, BRS Linda, Fantasya, Isabel, Isabel SM, Bordô, Marta SM, Concord Precoce, Niagara, Rubia, Cristal, VR 043-43, Paulsen 1103, Goethe, Catawba S, Catawba T, IAC 766, IAC 313, Gravesac, PESC 2, RR 101-14 Mgt, and SO4, most of them developed in Brazil.
The results of the present work corroborate the empirical and historical origin of grape germplasm in Santa Catarina. The recent expansion and renovation of viticulture in this state was based on the introduction of European germplasm, as shown by data from groups 1 and 2. Group 3, however, is predominantly composed of well-established and recently introduced American and hybrid grape varieties, which are still a valuable source of income for small farmers in Santa Catarina.
Conclusions
1. Genotype analyses using ten simple sequence repeat (SSR) loci was adequate to identify the grape germplasm in use in Santa Catarina.
2. The genotypes in these collections were mostly European varieties, but some American and hybrid varieties were also present.
3. The existence of unique regional germplasm was supported by the finding that several SSR genotypes did not match any molecular profile in the literature and databases.
4. Taken together the results can be used to better rationalize and manage grape collections, as well as for providing reference DNA profiles for the nursery industry and for breeding.
Acknowledgements
To the Progoethe Project, for financial support; to Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, for a fellowship; to the Autonomous Province of Trento, Universidade Federal de Santa Catarina, Empresa de Pesquisa Agropecuária e Extensão Rural de Santa Catarina, and Companhia Integrada de Desenvolvimento Agrícola de Santa Catarina; to José Vouillamoz (University of Neuchâtel, Switzerland), for helping in the analysis of molecular profiles.
Received on November 20, 2008 and accepted on April 30, 2009
- ADAM-BLONDON, A.F.; ROUX, C.; CLAUX, D.; BUTTERLIN, G.; MERDINOGLU, D.; THIS, P. Mapping 245 SSR markers on the Vitis vinifera genome: a tool for grape genetics. Theoretical and Applied Genetics, v.109, p.1017-1027, 2004.
- ANDRÉS, M.T. de; CABEZAS, J.A.; CERVERA, M.T.; BORREGO, J.; MARTÍNEZ-ZAPATER, J.M.; JOUVE, N. Molecular characterization of grapevine rootstocks maintained in germplasm collections. American Journal of Enology and Viticulture, v.58, p.75-86, 2007.
- BOWCOCK, A.M.; RUIZ-LINARES, A.; TOMFOHRDE, J.; MINCH, E.; KIDD, J.R.; CAVALLI-SFORZA, L.L. High resolution of human evolutionary trees with polymorphic microsatellites. Nature, v.368, p.455-457, 1994.
- BOWERS, J.E.; BOURSIQUOT, J.M.; THIS, P.; CHU, K.; JOHANSSON, H.; MEREDITH, C.P. Historical genetics: the parentage of Chardonnay, Gamay and other wine grapes of Northeastern France. Science, v.285, p.1562-1565, 1999a.
- BOWERS, J.E.; DANGL, G.S.; MEREDITH, C.P. Development and characterization of additional microsatellite DNA markers for grape. American Journal of Enology and Viticulture, v.50, p.243-246, 1999b.
- BOWERS, J.E.; DANGL, G.S.; VIGNANI, R.; MEREDITH, C.P. Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.). Genome, v.39, p.628-633, 1996.
- COSTANTINI, L.; MONACO, A.; VOUILLAMOZ, J.F.; FORLANI, M.; GRANDO, M.S. Genetic relationships among local Vitis vinifera cultivars from Campania (Italy). Vitis, v.44, p.25-34, 2005.
- DETTWEILER, E.; KRAUSE, D.J.; HARRER, S. The European Vitis database. 1998. Available at: <http://www.dainet.de/eccdb/vitis/>. Accessed on: 7 May 2009.
- DOYLE J.J.; DOYLE J.L. Isolation of plant DNA from fresh tissue. Focus, v.12, p.13-15, 1990.
- FELSENSTEIN, J. Phylogeny inference package. Available at: <http://evolution.genetics.washington.edu/phylip.html>. Accessed on: 7 May 2009.
- GRANDO, M.S.; COSTANTINI, L.; MADINI, A.; SEGALA, C.; CORRADINI, S. GMC, una collezione di profili molecolari di vite. L'Informatore Agrario, v.8, p.33-34, 2002.
- IBGE. Banco de dados agregados: relatório do ano de 2007.Available at: <http://www.sidra.ibge.gov.br>. Accessed on: 18 Aug. 2008.
- LEFORT, F.; ROUBELAKIS-ANGELAKIS, K.A. Genetic comparison of greek cultivars of Vitis vinifera L. by nuclear microsatellite profiling. American Journal of Enology and Viticulture, v.52, p.101-108, 2001.
- MARTÍN, J.P.; BORREGO, J.; CABELLO, F.; ORTIZ, J.M. Characterization of Spanish grapevine cultivar diversity using sequence-tagged microsatellite site markers. Genome, v.46, p.10-18, 2003.
- MINCH, E.; RUIZ-LINARES, A.; GOLDSTEIN, D.B.; FELDMAN, M.W.; CAVALLI-SFORZA, L.L. Microsat: a computer program for calculating various statistics on microsatellite allele data. Version 1.5. Stanford: Stanford University, 1997.
- PAGE, R.D.M. TreeView: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences, v.12, p.357-358, 1996.
- POLLEFEYS, P.; BOUSQUET, J. Molecular genetic diversity of the French-American grapevine hybrids cultivated in North America. Genome, v.46, p.1037-1048, 2003.
- RIAZ, S.; DANGL, G.S.; EDWARDS, K.J.; MEREDITH, C.P. A microsatellite marker based framework linkage map of Vitis vinifera L. Theoretical and Applied Genetics, v.108, p.864-872, 2004.
- SCHUCK, M.R.; MOREIRA, F.M.; VOLTOLINI, J.A.; GUERRA, M.P.; GRANDO, M.S.; SILVA, A.L. da. Studio genetico delle collezioni di vite nella provincia di Santa Catarina (Brasile) attraverso l'uso di marcatori microsatellite. In: CONGRESSO MONDIALE DELLA VIGNA E DEL VINO, 31.; ASSEMBLEA GENERALE DELL'O.I.V., 6., 2008, Verona. Riassunti delle comunicazioni. Verona: OIV: Ministero delle Politiche Agricole, Alimentari e Forestali, 2008. 1 CD-ROM.
- SEFC, K.M.; LEFORT, F.; GRANDO, M.S.; SCOTT, K.; STEINKELLNER, H.; THOMAS, M.R. Microsatellite markers for grapevine: a state of the art. In: ROUBELAKIS-ANGELAKIS, K.A. (Ed.). Molecular biology and biotechnology of grapevine. Amsterdam: Kluwer, 2001. p.433-463.
- SEFC, K.M.; LOPES, M.S.; LEFORT, F.; BOTTA, R.; ROUBELAKIS-ANGELAKIS, K.A.; IBAÑEZ, J.; PEJIC, I.; WAGNER, H.W.; GLOSSL, J.; STEINKELLNER, H. Microsatellite variability in grapevine cultivars from different European regions and evaluation of assignment testing to assess the geographic origin of cultivars. Theoretical and Applied Genetics, v.100, p.498-505, 2000.
- SEFC, K.M.; REGNER, F.; TURETSCHEK, E.; GLÖSSL, J.; STEINKELLNER, H. Identification of microsatellite sequences in Vitis riparia and their applicability for genotyping of different Vitis species. Genome, v.42, p.367-373, 1999.
- THIS, P.; JUNG, A.; BOCCACCI, P.; BORREGO, J.; BOTTA, R.; COSTANTINI, L.; CRESPAN, M.; DANGL, G.S.; EISENHELD, C.; FERREIRA-MONTEIRO, F.; GRANDO, S.; IBÁÑEZ, J.; LACOMBE, T.; LAUCOU, V.; MAGALHÃES, R.; MEREDITH, C.P.; MILANI, N.; PETERLUNGER, E.; REGNER, F.; ZULINI, L.; MAUL, E. Development of a standard set of microsatellite reference alleles for identification of grape cultivars. Theoretical and Applied Genetics, v.109, p.1448-1458, 2004.
- THOMAS, M.R.; SCOTT, N.S. Microsatellite repeats in grapevine reveal DNA polymorphisms when analysed as sequence-tagged sites (STSs). Theoretical and Applied Genetics, v.86, p.985-990, 1993.
- VIGNANI, R.; BOWERS, J.E.; MEREDITH, C.P. Microsatellite DNA polymorphism analysis of clones of Vitis vinifera 'Sangiovese'. Scientia Horticulturae, v.65, p.163-169, 1996.
- WAGNER, H.W.; SEFC, K.M. Identity 1.0: software for the analysis of microsatellite data. Vienna: University of Agricultural Sciences, 1999.
- WALKER, A.R.; LEE, E.; ROBINSON, S.P. Two new grape cultivars, bud sports of 'Cabernet Sauvignon' bearing pale-coloured berries, are the result of deletion of two regulatory genes of the berry colour locus. Plant Molecular Biology, v.62, p.623-635, 2006.
- ZULINI, L.; FABRO, E.; PETERLUNGER, E. Characterization of the grapevine cultivar Picolit by means of morphological descriptors and molecular markers. Vitis, v.44, p.35-38, 2005.
Publication Dates
-
Publication in this collection
17 July 2009 -
Date of issue
May 2009
History
-
Accepted
30 Apr 2009 -
Received
20 Nov 2008