Abstract
The present study aimed to analyze the genetic similarity between genomic profiles of thirteen Klebsiella oxytoca and seven Klebsiella pneumoniae samples isolated from two different collections carried out in different places of dental offices. Random amplified polymorphic DNA (RAPD) technique and similarity coefficients (calculated by Sorensen-Dice and simple matching) were applied to determine their genetic profile of randomic DNA sequences. The majority of the isolates of K. pneumoniae and K. oxytoca presented similar coefficient values (e" 0.80). Thus, it was possible to identify that strain dissemination occurred mainly via the hands of the surgeon-dentists and, finally, to determine the genetic similarity of the strains from dental office environments.
Klebsiella oxytoca; Klebsiella pneumoniae; RAPD; dental office
SHORT COMMUNICATION
Assessment of genetic relationship between Klebsiella pneumoniae and Klebsiella oxytoca samples isolated from a dental office
Pimenta-Rodrigues MVI, IV; Fusco-Almeida AMII; Bertoni BWI; Pietro RCLRII, III
IUnit of Medicinal Plant and Microorganism Biotechnology, Ribeirão Preto University, Ribeirão Preto, São Paulo State, Brazil
IIPharmaceutical Sciences Course, Ribeirão Preto University, Ribeirão Preto, São Paulo State, Brazil
IIISchool of Pharmaceutical Sciences, São Paulo State University, UNESP, Araraquara, São Paulo State, Brazil
IVDepartment of Tropical Diseases, Botucatu Medical School, São Paulo State University, UNESP, Botucatu, São Paulo State, Brazil
Correspondence to Correspondence to: Rosemeire Cristina Linhari Rodrigues Pietro Departamento de Fármacos e Medicamentos Faculdade de Ciências Farmacêuticas de Araraquara, UNESP Araraquara, SP, 14801-902, Brasil Phone: +55 16 3301 6965 Fax: +55 16 3301 6960 Email: pietrorc@fcfar.unesp.br
ABSTRACT
The present study aimed to analyze the genetic similarity between genomic profiles of thirteen Klebsiella oxytoca and seven Klebsiella pneumoniae samples isolated from two different collections carried out in different places of dental offices. Random amplified polymorphic DNA (RAPD) technique and similarity coefficients (calculated by Sorensen-Dice and simple matching) were applied to determine their genetic profile of randomic DNA sequences. The majority of the isolates of K. pneumoniae and K. oxytoca presented similar coefficient values (e" 0.80). Thus, it was possible to identify that strain dissemination occurred mainly via the hands of the surgeon-dentists and, finally, to determine the genetic similarity of the strains from dental office environments.
Key Words:Klebsiella oxytoca, Klebsiella pneumoniae, RAPD, dental office.
INTRODUCTION
Klebsiella pneumoniae and Klebsiella oxytoca are important opportunistic hospital-acquired pathogens that cause high morbidity and mortality rates among newborn infants, the elderly and immunocompromised patients. K. oxytoca cannot be affected by amino or carboxy penicillins; meanwhile, K. pneumoniae is highly resistant to broad-spectrum cephalosporins and aminoglycosides (4, 6). Outbreaks of these infectious species have been associated with a wide variety of sources and reservoirs, including lavatory basins and ultrasonographic gels (8, 18).
Hospital environments can be compared with dental offices, so that the control of emerging and persistent pathogenic microorganisms presents great importance (1). Both humidity and temperature of the oral cavity can create a wide range of habitats that present different environmental conditions and provide ideal media for microorganism colonization (8). Dental health professionals, due to their repeated exposure to these microorganisms, frequently are at high risk for developing infectious diseases (17).
Infection control efforts aim to identify the source of infection and the mode of transmission. Traditional techniques, based on phenotypic characteristics for classifying the Klebsiella genus, are often insufficiently sensitive for typing all strains and to discriminate among them (11). A distinctive polymorphism generated by the random amplified polymorphic DNA (RAPD) is a useful method for detecting strain differences and its ability to distinguish a wide variety of bacteria in a short time suggests that it constitutes a functional molecular epidemiological tool (2). Thus, the present study aimed to identify the transmission routes and genetic similarity of both microorganisms by the RAPD technique.
Thirteen Klebsiella oxytoca and seven Klebsiella pneumoniae samples from several sources (Table 1) were isolated and divided into two collections: the first was performed in April (Collection I) and the other in June (Collection II), both in 2006, within the Clinic of Surgery and Buco-Maxilo-Facial Traumatology, Ribeirão Preto University, São Paulo State, Brazil. The samples collected by conventional procedures employing swabs were submitted to growth in brain-heart infusion broth (BHI) (Biobrás, Brazil) directly plated on 5% sheep blood agar (Biobrás, Brazil) and MacConkey agar (Biobrás, Brazil), and were incubated under aerobic conditions for 24 hours at 37°C. The Klebsiella oxytoca and Klebsiella pneumoniae isolates were identified by the Probac Kit® (Probac do Brasil, Brazil) and maintained as frozen stocks at -70°C in the presence of 15% glycerol. The bacterial strains were cultured in BHI for 24 hours, at 37°C, and finally centrifuged for 30 minutes at 14,000 rpm (Centrifuge 5417R®, Eppendorf AG, Germany) for pellet formation for DNA extraction.
The extraction of genomic DNA was carried out using the GFX® Genomic Blood DNA Purification Kit (Amersham Pharmacia Biosciences Inc., UK) with modifications and adaptations to enzymatic buffers for cell lyses. For gram-negative strains, buffers employed were proteinase K (12 mM TRIS-HCl pH 8.0; 6 mM EDTA; 0.6% SDS) (Gibco-BRL, USA); and proteinase K (100 mg/mL in 10 mM, TRIS-HCl pH8.0) (Pharmacia Biotec, USA). The quantification of the genomic DNA was analyzed by estimating the band intensity in comparison with ë phage DNA (Promega Inc., USA) via 1% agarose gel electrophoresis. For RAPD analysis, preliminary assays were carried out to test 25 primers synthesized by Operon Technologies (USA) and Amersham Pharmacia Biosciences Inc. (UK). The primer Operon 18 (5'-CAGCACCCAC-3') was selected for the study on isolates of Klebsiella pneumoniae and Klesiella oxytoca, based on the accuracy and reproducibility of the amplified profiles. The polymerase chain reactions (PCR) had been prepared and optimized specifically for both bacterial isolates according to Pimenta-Rodrigues (13); in a 30-µL tube were added 3 µL of buffer 10x (Phoneutria Biotecnologia e Serviços Inc., Brazil), DNTPs (25 mM) and MgCl2 (25 mM) (Phoneutria Biotecnologia e Serviços Inc., Brazil); 5 µL of primers (10 ng/µL); 0.6 µL of Taq DNA polymerase (5 U/µL) (Phoneutria Biotecnologia e Serviços Inc., Brazil) and 3.0 µL genomic DNA (5 ng/µL). Amplification was performed in a thermocycler (PTC-100® Programmable Thermal Controller, MJ Researcher Inc., USA) programmed for two cycles of 2 minutes at 94°C, 1 minute at 37°C and 2 minutes at 72°C; followed by 33 cycles of 10 seconds at 94°C, 20 seconds at 40°C and 2 minutes at 72°C, and finally an amplification step of 5 minutes at 72°C. Amplification products were resolved by electrophoresis in a 2.0% agarose gel (Gibco/Invitrogen Amarillo, USA) in TBE buffer 1x (Tris 90 mM, boric acid 90 mM, EDTA 2 mM pH 8.0). The amplification products were stained with ethidium bromide and, thus, became visible under ultraviolet light, when they were photographed (Image Master® VDS, Pharmacia Biotech, USA). A 110 bp ladder (Amersham Pharmacia Biosciences Inc., UK) was included in each PCR run. The reproducibility of amplification results was evaluated in parallel experiments by three repetitions of the PCR reactions.
Similarity coefficients (Sab) were calculated based on absence and/or presence of bands. Dendrograms of Sab were generated using a binary code according to NTSYS program (Numerical Taxonomy and Multivariate Analysis System, version 2.1) (14). The Sab between the patterns for each pair of A and B strains was calculated by the formula Sab = 2E/(2E + a + b) using the Sorensen-Dice similarity coefficient that considers the joint absence of bands, and Sab = a + d/a + b + c + d utilizing the simple matching similarity coefficient that does not consider the joint absence of bands, where E is the number of common bands in A and B, a is the number of bands only in isolated A, b is the number of bands only in isolated B and d is the number of absent bands found in A and B isolates and also based on the Unweighted Pair Group Method with Arithmetic mean clustering method (UPGMA).
In the present study, the interpretation of these results was: for 1.0 similarity coefficient the isolates were designated genetically indistinguishable; for 0.99 to 0.80 coefficients, they were considered closely related (highly similar but not identical, however could be considered the same strain); for 0.79 to 0.50, isolates were possibly related; and for less than 0.50, they were considered unrelated (16). The averages established in the present study were performed according to Pfaller et al. (9); total Sab was estimated based on Sab values from the entire collection.
Of the total isolates, 50% were obtained from Collection I while 50% were from Collection II. And from the total, about 30% of K. oxytoca and about 20% of K. pneumoniae were found on the hand skins of surgeon-dentists and their gloves.
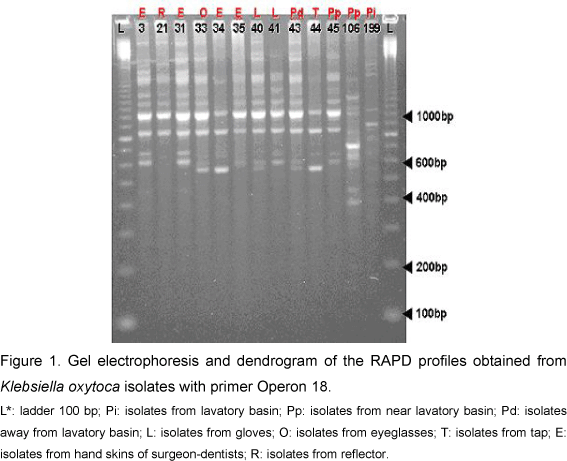
In each one of the thirteen K. oxytoca isolates, the PCR low accuracy permitted the amplification of 5 to 19 bands, ranging, in size, from 400 bp to about 2,000 bp (Figure 1). It was found that all isolates had a major band, about 1,000 bp. The isolates were grouped with a total Sab of about 0.89, so that the dendrogram analysis revealed two distinct groups (Figure 2). Group I contained ten isolates subdivided into two subgroups, Ia (nine isolates with Sab higher than 0.85) and Ib (only one isolate, from a lavatory basin, with Sab around 0.6). Different isolates from Subgroup Ia, similar to Sab 1.0, were derived from the skin, gloves and away from the lavatory basin. Group II, presenting three isolates from near the lavatory basin, tap and skin, had two isolates with coefficient Sab 1.0.
Results obtained from Klebsiella oxytoca samples by simple matching coefficient corroborated the division into groups I and II with Sab total average around 0.93 (Figure 3). Group I comprised twelve isolates subdivided into two subgroups, Ia (nine isolates with Sab varying between 0.86 and 1.0) and Ib (three isolates from the lavatory basin, tap and skin, including two isolates with 1.0 Sab coefficient). Group II had only one isolate from the collection near the lavatory basin, with Sab around 0.58.
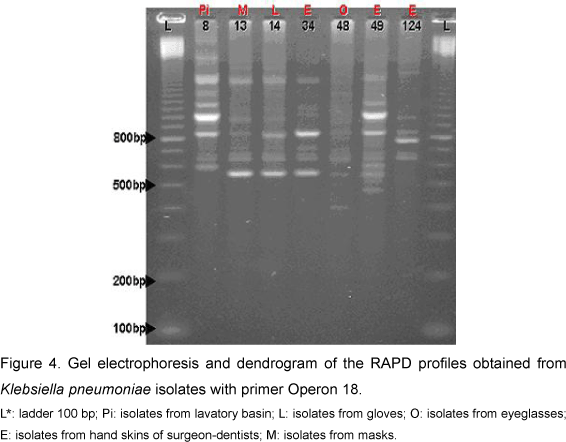
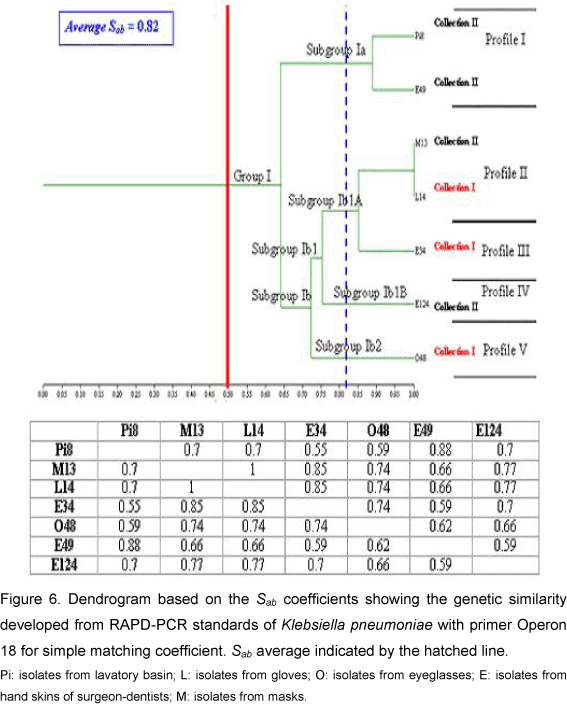
For Klebsiella pneumoniae the PCR reaction permitted the amplification of 9 to 16 bands ranging in size from 350 bp to 2,000 bp (Figure 4). The resultant dendrogram of the seven bacterial isolates, by the RAPD analysis, presented total Sab around 0.79. All the isolates were aggregated into a single group and subdivided into two subgroups (Figure 5). Subgroup Ia presented two isolates (from hand skins of dentists and lavatory basin) that presented similarity coefficient around 0.90. Subgroup Ib was subdivided into two other subgroups: Ib1 and Ib2. In Ib2 only one isolate - from hand skins of dentists - was similar to the Subgroup Ib1, with Sab around 0.67. The Subgroup Ib1 was further divided into Ib1A and Ib1B; one isolate of Ib1B was collected from eyeglasses and presented similarity coefficient near 0.70. Subgroup Ib1A consisted of two isolates with Sab 1.0, originating from masks and gloves, and an additional isolate with similarity coefficient around 0.87 (on dentists' hands). The results from the simple matching coefficient, with Sab total average 0.82, were similar to the Sorensen-Dice coefficient, although the difference was coefficient values that were higher for simple matching (Figure 6).
The opportunistic pathogen Klebsiella ssp usually originate in immunocompromised individuals that are hospitalized and carry underlying illnesses such as diabetes mellitus or chronic pulmonary obstruction. The principal reservoirs of Klebsiella ssp are the gastrointestinal tract and the hands of hospital personnel (7). Based on a simple technique, RAPD is viable for large-scale use in epidemiological studies, in association with bacterial samples that help to identify possible origins of contamination and infection. This method is sufficiently sensitive when parameters of reaction optimization and choice of primers are applied adequately (5, 12, 15, 19).
In the present study the isolate similarity profile was evaluated by two methods. In general terms, it was observed that, although the simple matching coefficient considers band joint absences, this method did not present considerable differences in relation to the Sorensen-Dice coefficient, which does not consider joint absence of bands. This fact can be explained on the basis that joint absences do not necessarily mean that DNA regions are identical, which suggests the use of any coefficient that does not consider the joint absence, as does Sorensen-Dice.
The phylogenetic analysis generated by the dendrograms enabled the characterization of K. oxytoca individuals. Through evaluation of Sab values, it can be concluded that all isolates belonging to Ia, except for the sample Pi199, were the same strain; therefore, they present Sab higher than 0.80 in relation to the isolates with Sab 1.0. These isolates were collected from skin, eyeglasses, gloves, reflectors and places near lavatory basins, which suggests dissemination by hands. Additionally, as these isolates were present in different collections, we believe that this bacterial strain persisted in the clinical environment.
The examination of K. pneumoniae by RAPD permitted - through observation of Sab values in Subgroup Ia - detection of the presence of two isolates, with Sab 0.90, collected from lavatory basins and hand skins. This similarity coefficient value showed that these isolates were part of the same strain. Meanwhile, in Subgroup Ib1A that contained three isolates, two presented Sab 1.0 and were derived from masks and gloves; and the other, with Sab around 0.81, was from hand skins. These data imply dissemination by hands, since they present high similarity among isolates found on hand skins of dentists. These strains are supposed to be prevalent in clinical environments, since isolates from gloves were obtained in Collection I and those derived from masks, accquired in Collection II, present Sab 1.0. Eisen et al. (3), through the phylogenetic analysis of K. pneumoniae isolates, found that handling (by medical staff) in newborn units was the usual non-environmental source of dissemination for these microorganisms.
Due to the close connection among these isolates, another hypothesis suggested that instead of being contaminated by infected hands, the environment itself could disseminate microorganisms to hands, gloves and masks. This supposition agrees with Podschun et al. (10) who demonstrated the incidence of Klebsiella species on surface waters and proved its ability to express virulence factors, thus evidencing the importance of the presence of these species in clinical environments. In this case, both dissemination routes of closely related pathogenic strains offer risks of cross-contamination.
Knowledge of the genetic interactions among species of the genus Klebsiella provides a framework for studies on the distribution of phenotypic properties implicated in virulence or in epidemiological differences between clones (2). RAPD studies would be useful to determine whether the K. pneumoniae and K. oxytoca clusters correspond to distinct genomic species and given the proof that it is a useful technique to distinguish between related and unrelated isolates of K. pneumoniae and K. oxytoca. The capacity to detect genetic heterogeneity in different strains is important for the surveillance of odontological-acquired infections and may be used in epidemiologic studies, in association with strains that help identify the origin of contamination and infections.
ACKNOWLEDGEMENTS
We are grateful to the PROSUP/CAPES program for the M.Sc. scholarship of Marcus Vinicius Pimenta Rodrigues and to the financial support of Ribeirão Preto University, UNAERP.
Received: December 4, 2007
Accepted: August 20, 2008
Abstract published online: September 1, 2008
Full paper published online: November 30, 2008
Conflicts of interest: There is no conflict.
- 1 Bernardo WLC., Boriollo MFG., Gonçalves RB., Hofling JF. Staphylococcus aureus ampicillin-resistant from the odontological clinic environment. Rev. Inst. Med. Trop. São Paulo, 2005, 47, 19-24.
- 2 BRISSE S., VERHOEF J. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int. J. Syst. Evol. Microbiol, 2001, 51, 915-24.
- 3 EISEN D., RUSSEL EG., TYMMS M., ROPER EJ., GRAYSON ML., TURNIDGE J. Random amplified polymorphic DNA and plasmid analyses used in investigation of an outbreak of multiresistant Klebsiella pneumoniae. J. Clin. Microbiol., 1995, 33, 713-7.
- 4 FEVRE C., JBEL M., PASSET V., WEILL FX., GRIMONT PAD., BRISSE S. Six groups of the OXY β-lactamase evolved over millions of years in Klebsiella oxytoca Antimicrob. Agents Chemother, 2005, 49, 3453-62.
- 5 HILTON AC., BANKS JG., PENN CW. Optimization of RAPD for fingerprinting Salmonella Lett. Appl. Microbiol., 1997, 24, 243-8.
- 6 KURUPATI P., CHOW C., KUMARASINGHE G., POH CL. Rapid detection of Klebsiella pneumoniae from blood culture bottles by real-time PCR. J. Clin. Microbiol, 2004, 42, 1337-40.
- 7 LINCOPAN N., MCCULLOCH JA., REINERT C., CASSETARI VC., GALES AC., MAMIZUKA EM. First isolation of metallo-β-lactamase-producing multiresistant Klebsiella pneumoniae from a patient in Brazil. J. Clin. Microbiol, 2005, 43, 516- 9.
- 8 MONARCA S., GROTTOLO M., RENZI D., PAGANELLI C., SAPELLI P., ZERBINI I., NARDI G. Evaluation of environmental bacterial contamination and procedures to control cross infection in a sample of Italian dental surgeries. Occup. Environ. Med., 2000, 57, 721-6.
- 9 PFALLER MA., LOCKHART SR., PUJOL C., SWAILS-WENGER JA., MESSER SA., EDMOND MB., JONES RN., WENZEL RP., SOLL DR. Hospital specificity, region specificity, and fluconazole resistance of Candida albicans bloodstream isolates. J. Clin. Microbiol, 1998, 36, 1518-29.
- 10 PODSCHUN R., PIETSCH S., HOLLER C., ULLMANN U. Incidence of Klebsiella species in surface waters and their expression of virulence factors. Appl. Environ. Microbiol., 2001, 67, 3325-7.
- 11 PODSCHUN R., ULLMANN U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev, 1998, 11, 589-603.
- 12 QUINTAES BR., LEAL NC., REIS EM., HOFER E. Optimization of randomly amplified polymorphic DNA-polymerase chain reaction for molecular typing of Salmonella enterica serovar typhi. Rev. Soc. Bras. Med. Trop, 2004, 37, 143-7.
- 13 RODRIGUES MVP. Molecular typing of bacterial strains isolated from the clinics of surgery and face traumatology at Ribeirão Preto University. J. Venom. Anim. Toxins incl. Trop. Dis, 2007, 13, 894.
- 14 ROLHF JF. NTSYS - pc: numerical taxonomy and multivariate analysis system. Version 2.1. Setauket. NY: Exeter Software, 2000. 38p.
- 15 SANTOS LR. Standardization of RAPD (Random Amplification of Polymorphic DNA) for fingerprinting of Salmonella enteritidis Rev. Fac. Zootec. Vet. Agron., 2003, 10, 144-58.
- 16 Tenover FC., Arbeit RD., Goering RV., Mickelsen PA., Murray BE., Persing DH., Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin Microbiol., 1995, 33, 2233-9.
- 17 Toroðlu SM., Haytaç MC., Köksal F. Evaluation of aerosol contamination during debonding procedures. Angle Orthod., 2001, 71, 299-306.
- 18 VAN'T VEEN A., VAN DER ZEE A., NELSON J., SPEELBERG B., KLUYTMANS JAJW., BUITING AGM. Outbreak of infection with a multiresistant Klebsiella pneumoniae strain associated with contaminated roll boards in operating rooms. J. Clin. Microbiol., 2005, 43, 4961-7.
- 19 YU K., PAULS KP. Optimization of the PCR program for RAPD analysis. Nucleic Acids Res, 1992, 20, 2606.
Publication Dates
-
Publication in this collection
09 Dec 2008 -
Date of issue
2008
History
-
Received
04 Dec 2007 -
Accepted
20 Aug 2008