Abstracts
This work aimed to assess pathogenic potential and clonal relatedness of Aeromonas sp. and Vibrio cholerae isolates recovered during a diarrhea outbreak in Brazil. Clinical and environmental isolates were investigated for the presence of known pathogenic genes and clonal relatedness was assessed by intergenic spacer region (ISR) 16S-23S amplification. Four Aeromonas genes (lip, exu, gcat, flaA/B) were found at high overall frequency in both clinical and environmental isolates although the lip gene was specifically absent from selected species. A fifth gene, aerA, was rarely found in A. caviae, the most abundant species. The ISR profile revealed high heterogeneity among the Aeromonas isolates and no correlation with species identification. In contrast, in all the V. cholerae isolates the four genes investigated (ctxA, tcpA, zot and ace) were amplified and revealed homogeneous ISR and RAPD profiles. Although Aeromonas isolates were the major enteric pathogen recovered, their ISR profiles are not compatible with a unique cause for the diarrhea events, while the clonal relationship clearly implicates V. cholerae in those cases from which it was isolated. These results reinforce the need for a better definition of the role of aeromonads in diarrhea and whether they benefit from co-infection with V. cholerae.
Aeromonas; Vibrio cholerae; Diarrhea; Virulence; PCR; Pathogenicity
O objetivo deste trabalho foi estabelecer o potencial patogênico e a relação clonal de isolados de Aeromonas sp. e Vibrio cholerae obtidos durante um surto de diarréia. Isolados clínicos e ambientais foram investigados quanto à presença de genes de virulência e sua relação clonal foi obtida através de amplificação da Região Espaçadora Intergênica (REI) 16S-23S. Quatro genes de Aeromonas (lip, exu, gcat, flaA/B) foram encontrados em alta frequência embora o gene lip tenha se mostrado ausente em algumas espécies. Um quinto gene, aerA, foi raramente encontrado em A. caviae, a espécie mais abundante. O perfil da REI revelou alta heterogeneidade entre os isolados de Aeromonas e nenhuma correlação com espécie. Em contraste, todas as amostras de V. cholerae amplificaram os genes investigados (ctxA, tcpA, zot e ace) e revelaram perfil clonal através de REI e RAPD. Embora Aeromonas tenha sido o principal patógeno isolado, o perfil da REI não é compatível como única causa para os eventos de diarréia, enquanto a relação clonal de V. cholerae aponta esse microrganismo como o provável agente do surto. Estes resultados reforçam a necessidade de definir melhor o papel de Aeromonas em diarréias e de que forma essas bactérias se beneficiam quando em co-infecção com V. cholerae.
MICROBIOLOGY
Molecular characterization of Aeromonas spp. and Vibrio cholerae O1 isolated during a diarrhea outbreak
Caracterização molecular de Aeromonas spp. e Vibrio cholerae O1 isolados durante um surto de diarréia
Carina Lucena Mendes-MarquesI; Larissa Mélo do NascimentoI; Grace Nazareth Diogo TheophiloII; Ernesto HoferII; Osvaldo Pompílio de Melo NetoI; Nilma Cintra LealI
IDepartamento de Microbiologia. Centro de Pesquisas Aggeu Magalhães/Fiocruz. Av. Professor Moraes Rego s/n, Cidade Universitária, 50670-420 Recife, PE, Brazil, Tel. +55.81.21012568
IIDepartamento de Bacteriologia, Instituto Oswaldo Cruz/Fiocruz, Av. Brasil 4365, Manguinhos, 21040-360 Rio de Janeiro, RJ, Brazil, Tel. +55.21.25984220
Correspondence Correspondence to: Carina Lucena Mendes-Marques. Centro de Pesquisas Aggeu Magalhães/Fiocruz, Departamento de Microbiologia. Av. Prof. Moraes Rego s/n, Cidade Universitária, 50670-420 Recife, PE, Brasil. Tel.: +55.81.21012568. Fax: +55.81.21012647. E-mail: clmendes@gmail.com
SUMMARY
This work aimed to assess pathogenic potential and clonal relatedness of Aeromonas sp. and Vibrio cholerae isolates recovered during a diarrhea outbreak in Brazil. Clinical and environmental isolates were investigated for the presence of known pathogenic genes and clonal relatedness was assessed by intergenic spacer region (ISR) 16S-23S amplification. Four Aeromonas genes (lip, exu, gcat, flaA/B) were found at high overall frequency in both clinical and environmental isolates although the lip gene was specifically absent from selected species. A fifth gene, aerA, was rarely found in A. caviae, the most abundant species. The ISR profile revealed high heterogeneity among the Aeromonas isolates and no correlation with species identification. In contrast, in all the V. cholerae isolates the four genes investigated (ctxA, tcpA, zot and ace) were amplified and revealed homogeneous ISR and RAPD profiles. Although Aeromonas isolates were the major enteric pathogen recovered, their ISR profiles are not compatible with a unique cause for the diarrhea events, while the clonal relationship clearly implicates V. cholerae in those cases from which it was isolated. These results reinforce the need for a better definition of the role of aeromonads in diarrhea and whether they benefit from co-infection with V. cholerae.
Keywords:Aeromonas;Vibrio cholerae; Diarrhea; Virulence; PCR; Pathogenicity.
RESUMO
O objetivo deste trabalho foi estabelecer o potencial patogênico e a relação clonal de isolados de Aeromonas sp. e Vibrio cholerae obtidos durante um surto de diarréia. Isolados clínicos e ambientais foram investigados quanto à presença de genes de virulência e sua relação clonal foi obtida através de amplificação da Região Espaçadora Intergênica (REI) 16S-23S. Quatro genes de Aeromonas (lip, exu, gcat, flaA/B) foram encontrados em alta frequência embora o gene lip tenha se mostrado ausente em algumas espécies. Um quinto gene, aerA, foi raramente encontrado em A. caviae, a espécie mais abundante. O perfil da REI revelou alta heterogeneidade entre os isolados de Aeromonas e nenhuma correlação com espécie. Em contraste, todas as amostras de V. cholerae amplificaram os genes investigados (ctxA, tcpA, zot e ace) e revelaram perfil clonal através de REI e RAPD. Embora Aeromonas tenha sido o principal patógeno isolado, o perfil da REI não é compatível como única causa para os eventos de diarréia, enquanto a relação clonal de V. cholerae aponta esse microrganismo como o provável agente do surto. Estes resultados reforçam a necessidade de definir melhor o papel de Aeromonas em diarréias e de que forma essas bactérias se beneficiam quando em co-infecção com V. cholerae.
INTRODUCTION
Aeromonas are gram-negative bacilli from the Aeromonadaceae family found in aquatic environments: rivers and lakes and in both treated and raw sewage. They are known to be pathogenic to poikilothermic animals, causing ulcerative infections, and recently have been found to be associated with a variety of human extra-intestinal infections17. Their role as diarrhea causing agents is still controversial12 as some studies have found aeromonads carried as transient flora in healthy asymptomatic individuals2,15,17.
Although Aeromonas spp. produces virulence factors similar to other human enteropathogens4, and despite the fact that there are some studies with mutant strains where lack of specific virulence factors may be associated with loss of pathogenicity6,29, there is no animal model that reproduces the diarrhea so that the identification of virulence factors essential for its pathogenicity is therefore impaired. Hence, the role of Aeromonas in enteric infections have been mostly defined based on case reports, case-control studies and outbreak investigations associated with findings of the bacteria in diarrheic stools19, thus linking the microorganism to the disease3,17, although it has been shown that pathogenic aeromonads induces active Cl- secretion in the intestinal epithelium10.
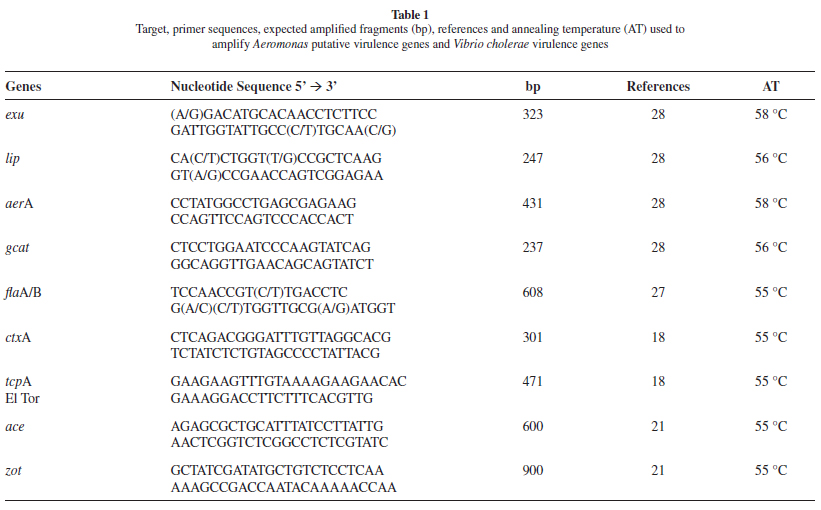
The Aeromonas' genomes harbor genes coding for putative virulence factors needed for different stages of infection, such as invasion, colonization and proliferation11: the flaA/B genes, coding for the polar flagellum, involved in the adhesion process which is essential for epithelium colonization22; the exu gene, coding for an extracellular DNase which blocks the antibacterial host defenses5; lip and gcat genes coding for extracellular lipases which make host cells membrane more susceptible to other toxins26; and the aerA gene, the most studied Aeromonas virulence factor, coding for a toxin, aerolysin, that induces pore formation in host cells membranes17.
In 2004, a diarrhea outbreak occurred in São Bento do Una, Pernambuco, Brazil, with 2,170 cases and an attack rate of 4.65%. At that occasion, the supply of drinking water was precarious and people used water without microbiological control16 suggesting that the water was the vehicle of infection. During the outbreak, 582 stool samples were analyzed and an enteric pathogen was isolated from 145 of these. Aeromonas spp. were the most frequent sole pathogen isolated from stools of patients suffering acute diarrhea (identified from 119 of the 145 samples where an enteric pathogen was recovered), with Vibrio cholerae O1 isolates also being recovered from a substantially smaller number of strains (18 out of 145)16. V. cholerae O1, the agent of cholera disease, is also a natural inhabitant of aquatic environments whose pathogenicity mechanisms are well defined24. Here, in an attempt to better characterize the pathogenic potential of the Aeromonas isolates, we have investigated the presence of putative virulence genes and assessed their clonal relatedness through ISR 16S-23S PCR analysis, performing parallel experiments with the V. cholerae isolates recovered during the same outbreak.
MATERIAL AND METHODS
Bacterial strains: One hundred and six Aeromonas spp. strains, out of a total of 119 originally isolated, and 18 V. cholerae O1 isolates from diarrheic stools and 19 Aeromonas spp. and seven V. cholerae isolates from aquatic environments were analyzed. Eighty two Aeromonas strains were kindly identified to the species level through 16S restriction fragment length polymorphism (RFLP) by Dr. Maria Jose Figueras from University Rovira i Virgili, Tarragona, Spain. Forty three strains were classified to the genus level, using biochemical tests1, and are collectively called Aeromonas sp in this work. All V. cholerae O1 strains were identified by biochemical25 and serological tests30. The differentiation between Vibrio and Aeromonas isolates was performed as previously described13. The reference strains Aeromonas hydrophila ATCC 7966T, Aeromonas veronii biotype veronii ATCC 35624T, Aeromonas caviae ATCC 15468T, and V. cholerae 569B T were included as controls. The cultures were stored at -80 ºC in BHI plus 25% glycerol. DNA from the various strains was obtained as previously described21.
PCR reactions: The presence of confirmed or putative virulence genes in the Aeromonas or Vibrio strains was assessed by PCR using primers described in the literature (Table 1). The lip, exu, aerA, gcat and flaA/B genes were investigated from the Aeromonas strains with the cholera toxin (ctxA), toxin co-regulated pilus subunit A (tcpA), accessory cholera enterotoxin (ace) and zonula occludens toxin (zot) genes being investigated from V. cholerae. The Aeromonas strains were also tested for ctxA to exclude the possibility of CTXΦ phage horizontal transfer between V. cholerae and Aeromonas strains. The Intergenic Spacer Region (ISR) between the 16S and 23S rDNA genes was amplified as previously described9. RAPD was performed with the primer CCGCAGCCAA as previously described21. PCR reactions were carried out in a Biometra T-3000 Genetic Analyzer thermal cycler using standard procedures and optimal annealing temperatures specific for each primer pair. PCR products were submitted to electrophoresis in agarose gels containing SYBR Safe DNA gel stain (Invitrogen) and photographed using the Kodak 1D Image Analysis software, version 3.5 (Digital Kodak Science).
Identification of PCR products: To confirm the identity of the amplified Aeromonas fragments, PCR products from one clinical (A. hydrophila ATCC 7966T) and one environmental (A. caviae) isolates were sequenced on ABI Prism 3100 Genetic Analyzer, Applied Biosystems, Foster City, CA, using the same PCR primers. Sequences were aligned using the BLASTn program. The identity of the amplified Vibrio PCR gene fragments was confirmed by comparing their sizes with equivalent fragments predicted from the V. cholerae 569B reference strain.
Statistics: A chi-square or Fisher's exact test was used to compare the virulence gene frequencies from clinical and environmental isolates. All conclusions are based on 5% significance level. The softwares Excel 2000 and R v2.10 were used.
RESULTS
Aeromonas species identification: From a total of 125 (106 clinical and 19 environmental) Aeromonas isolates recovered during the 2004 diarrhea outbreak and available for this study, 57 were identified as A. caviae (51 clinical/6 environmental isolates, respectively), 13 were A. veronii (12/1), four were A. hydrophila (2/2), four were A. media (clinical isolates only), three were A. trota (also clinical isolates), one clinical isolate was A. jandaei and 43 (33/10) were identified only to the genus level and classified as Aeromonas sp (these represent the strains which could not be identified by RFLP, considered the gold standard method to Aeromonas identification10,15). A. caviae and A. veronii isolates comprise then 85% (70 of 82) of those classified to the species level.
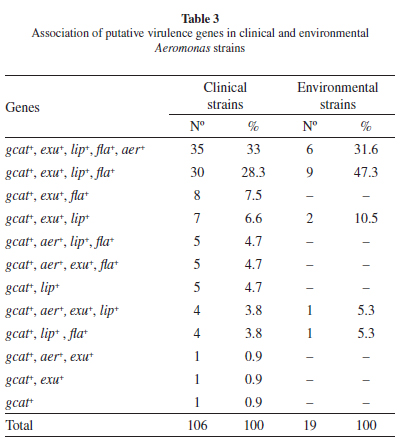
Virulence gene frequenciesfor Aeromonas spp isolates: To compare the pathogenic potential of the different Aeromonas isolates, the presence of five different putative virulence genes was investigated (lip, exu, gcat, flaA/B and aerA) through conventional PCR amplification. Amplified fragments were recovered for all five genes from multiple isolates and their identity was confirmed through the sequencing of representative fragments, from one clinical and one environmental isolates for each gene, and the alignment of the resulting sequences with the corresponding genes deposited in the GenBank database. When all clinical and environmental isolates were considered, the fragment corresponding to the gcat gene was the only one amplified from all strains; the flaA/B genes was amplified from 87 (82.1%) of the clinical and 16 (84.2%) of the environmental strains; the lip gene from 90 (84.9%) of the clinical and 19 (100%) of the environmental strains; the exu gene from 91 (85.8%) of the clinical and 18 (94.7%) of the environmental strains, and the aerA gene was generated in 50 (47.2%) of the clinical and seven (36.8%) of the environmental strains (Table 2). The overall gene frequencies between clinical and environmental isolates were statistically similar (p > 0.05). All five genes were amplified in 33% of the clinical and 31.6% of the environmental Aeromonas strains and 60.3% of the clinical and 68.4% of the environmental strains amplified at least three genes (Table 3).
When the gene frequencies were evaluated only for those isolates identified to the species level, marked differences in frequency for the lip and aerA genes were observed between species. The lip gene, present in all environmental strains, was also found in most A. caviae (45 of 57) and in all A. veronii and A. media clinical isolates, however it was not detected in any of the A. trota or the A. jandaei isolates. In contrast, the aerA gene was found in only 15 of the 57 A. caviae isolates, despite being present in most of the isolates from the remaining species (20/25). Indeed the five genes were amplified from all four A. media and 10 of the 13 A. veronii isolates but only from 11 of the 57 A. caviae strains (Table 2).
ISR 16S-23S profiling for Aeromonas spp isolates: To evaluate the genetic relatedness of the various isolates and compare how these varies within and between species, and also within those classified only as Aeromonas sp., the amplification of the ISR 16S-23S from all 125 isolates was carried out. Nine different profiles were obtained (Fig. 1), herein called R1 to R9, all containing a common band, approximately 550 bp, plus others which varied according to each profile. Most of the strains (57.6%) fitted into the profile R1, 13.6% into R7, 8% into R5, 4.8% into both R8 and R4, 4% into R3, 3.2% into R2, 2.4% into R6 and 1.6% into R9. When the different profiles were compared to those isolates classified at the species level, no specific profile could be definitively linked to any particular species. Nine of the 13 A. veronii isolates fitted into the R7 profile, whilst 39 of the 52 A. caviae isolates were classified within R1, but no single profile was found in only one species and no species, with the exception of A. jandaei which was represented by only one isolate, was represented by only one profile.
Virulence gene frequency, RAPD and ISR 16S-23S profiling for V. cholerae isolates: To evaluate the virulence potential of the 18 clinical and seven environmental Vibrio cholerae isolates recovered during the same outbreak, all identified as V. cholerae O1 Ogawa, the presence of the known virulence genes ctxA, tcpA, ace and zot (for details see Material and Methods) was investigated through PCR and found in all isolates tested. Next, genetic relatedness was investigated through ISR 16S-23S amplification and, contrary to what was observed for the Aeromonas strains, even those from a single species, all V. cholerae O1 strains fitted into a single ISR 16S-23S profile (Fig. 2). All V. cholerae O1 isolates from the outbreak region showed the same RAPD profile observed in two strains from the 1993 outbreak, when cholera entered Brazil, suggesting the persistence of a clone in the environment. Only a non-toxigenic environmental strain from a second geographical region without human cases showed slight differences in profile when this technique was performed (data not shown).
DISCUSSION
In spite of the high frequency of potentially virulent Aeromonas isolates in patient's feces during the evaluated diarrheal cases, the multiple species identified with distinct repertoires of virulence genes and heterogeneity in ISR 16S-23S sequences are not compatible with a single or related Aeromonas strain being responsible for the outbreak. On the other hand the Vibrio isolates analyzed revealed a homogeneous ISR and RAPD profile and high pathogenic potential associated with the presence of all searched virulence genes, clearly implicate V. cholerae as the etiological agent for those infections where it was found. Although it is not possible to rule out that the whole outbreak was due to an increased exposure of the target population to multiple enteropathogens, the possibility remains that some, if not most, of the cases where only Aeromonas strains were also isolated were also due to V. cholerae. It is possible that Aeromonas could be present as part of the patients' transient enteric flora and competed with Vibrio in vitro in the culture media, masking its presence and so, the real etiology of the disease. Alternatively, a related explanation would be for the Aeromonas strain to succeed a preliminary Vibrio infection in patients debilitated by the primary event.
Although Aeromonas could not be recognized as the etiological agent of the diarrhea event in São Bento do Una, the high frequency of putative virulence genes suggest its pathogenic potential. The overall similarity of ISR 16S-23S profiles and frequency of virulence genes among clinical and environmental strains suggests environmental contamination by infected people feces and probably from animal carriers due to inadequate sanitation in that city. Various water environments constitute Aeromonas ecological niches17 from where different bacterial lineages could spread to the city's inhabitants and proliferate, at least in immunocompromised individuals11.
The gene gcat, which codes for a lipase that modifies the host cells permeability and raises its accessibility to toxins, was present in all Aeromonas strains investigated, regardless of their origin, confirming what was previously described and that this gene represents a marker to distinguish Aeromonas from other enteropathogens 7,8. High frequency of gcat was also reported in another study8 and in our analysis, despite the different species and genetic background, it was consistently amplified from all Aeromonas strains assayed. Hence, the presence of this gene could represent a marker to distinguish Aeromonas from other enteropathogens7. The lip and exu genes code for antibacterial host defense factors and were also detected at high frequency in the strains analyzed. For the lip gene, its absence from selected species could be due to a failure of the amplification reaction related to its polymorphic nature, however degenerate primer pairs were used for these reactions specifically to maximize gene amplification in cases of polymorphisms. It is also possible that lack of amplification may be a consequence of the small number of strains investigated for A. trota and A. jandaei. Nevertheless, considering the high frequency observed for the lip gene in strains from the remaining species (frequency rates varying from 50 to 100%) we are confident that this observation may reflect a real difference in virulence gene profile and which should further investigated in the future. Lipases also play a role on bacterial nutrition26 and in the present study, the absence of the lip gene from clinical isolates of selected species, and its universal presence in the environmental isolates, may reflect more a role for survival in extracellular environment than in pathogenesis. The flaA/B genes essential for adhesion and epithelium colonization were found at high frequency in both clinical and environmental isolates. Aeromonas ability to form biofilms is directly related to the presence of the polar flagellum20. Therefore, the presence of flaA/B gene could be a virulence marker for Aeromonas.
Some authors associate the presence of high number of virulence genes with a higher pathogenic potential among Aeromonas strains14,23. A. hydrophila and A. veronii bv sobria showed a higher virulence potential compared to A. caviae14. Here, the frequency of aerA gene was indeed lower in A. caviae when compared with the remaining species, but still A. caviae was by far the most common species isolated from patients suggesting that this species remains virulent even in the absence of the aerolysin gene. Although the role of Aeromonas in diarrhea is not yet defined17, the incidence of these bacteria in feces of patients with diarrhea has significantly increased and, in agreement with the concern raised with regards to Aeromonas within the scientific community, we therefore recommend the routine investigation for these bacteria in all coprocultures.
ACKNOWLEDGEMENTS
The authors are thankful to Dr. Maria Jose Figueras (University Rovira i Virgili, Tarragona, Spain) for the RFLP analysis and to the Program for Technological Development in Tools for Health-PDTIS-Fiocruz for the use of its facilities.
Received: 9 January 2012
Accepted: 5 June 2012
- 1. Abbott SL, Cheung KW, Janda JM. The Genus Aeromonas: biochemical characteristics, atypical reactions, and phenotypic identification schemes. J Clin Microbiol. 2003;41:2348-57.
- 2. Albert MJ, Ansaruzzaman M, Talukder KA, Chopra AK, Kuhn I, Rahman M, et al Prevalence of enterotoxin genes in Aeromonas spp. isolated from children with diarrhea, healthy controls, and the environment. J Clin Microbiol. 2000;38:3785-90.
- 3. Altwegg M, Geiss HK. Aeromonas as a human pathogen. CRC Crit Rev Microbiol. 1989;16:253-86.
- 4. Borchardt MA, Stemper ME, Standridge JH. Aeromonas isolates from human diarrheic stool and groundwater compared by pulsed-field gel electrophoresis. Emerg Infect Dis. 2003;9:224-8.
- 5. Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532-5.
- 6. Burr SE, Pugovkin D, Wahli T, Segner H, Frey J. Attenuated virulence of an Aeromonas salmonicida subsp. salmonicida type III secretion mutant in a rainbow trout model. Microbiology. 2005;151(Pt 6):2111-8.
- 7. Chacón MR, Castro-Escarpulli G, Soler L, Guarro JA, Figueras MJ. DNA probe specific for Aeromonas colonies. Diagn Microbiol Infect Dis. 2002;44:221-5.
- 8. Chacón MR, Figueras MJ, Castro-Escarpulli G, Soler L, Guarro JA. Distribution of virulence genes in clinical and environmental isolates of Aeromonas spp. Antonie Van Leeuwenhoek. 2003;84:269-78.
- 9. Chun J, Huq A, Colwell RR. Analysis of 16S-23S rRNA intergenic spacer regions of Vibrio cholerae and Vibrio mimicus Appl Environ Microbiol. 1999;65:2202-8.
- 10. Epple HJ, Mankertz J, Ignatius R, Liesenfeld O, Fromm M, Zeitz M, et al. Aeromonas hydrophila beta-hemolysin induces active chloride secretion in colon epithelial cells (HT-29/B6). Infect Immun. 2004;72:4848-58.
- 11. Figueras MJ. Clinical relevance of Aeromonas sM503. Rev Med Microbiol. 2005;16:145-53.
- 12. Figueras MJ, Horneman AJ, Martinez-Murcia A, Guarro J. Controversial data on the association of Aeromonas with diarrhoea in a recent Hong Kong study. J Med Microbiol. 2007;56:996-8.
- 13. Ghenghesh KS, Ahmed SF, El-Khalek RA, Al-Gendy A, Klena J. Aeromonas-associated infections in developing countries. J Infect Dev Ctries. 2008;2:81-98.
- 14. Guerra IMF, Fadanelli R, Figueiró M, Schreiner F, Delamare APL, Wollheim, C et al Aeromonas associated diarrhoeal disease in south Brazil: prevalence, virulence factors and antimicrobial resistance. Braz J Microbiol. 2007;38:638-43.
- 15. Hanninen ML, Salmi S, Mattila L, Taipalinen R, Siitonen A. Association of Aeromonas spp. with travellers' diarrhoea in Finland. J Med Microbiol. 1995;42:26-31.
- 16. Hofer E, Reis CMF, Theophilo GND, Cavalcanti VO, Lima NV, Henriques MFCM. Envolvimento de Aeromonas em surto de doença diarréica aguda em São Bento do Una, Pernambuco. Rev Soc Bras Med Trop. 2006;39:217-20.
- 17. Janda JM, Abbott SL. The genus Aeromonas: taxonomy, pathogenicity and infection. Clin Microbiol Rev. 2010;23:35-73.
- 18. Keasler SP, Hall RH. Detecting and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet. 1993;341:1661.
- 19. Kirov SM. Aeromonas and Plesiomonas In: Doyle MP, Beuchat LR, Montville TJ, editors. Food Microbiology: fundamentals and frontiers. Washington, DC: ASM Press; 2001. p. 301-27.
- 20. Kirov SM, Castrisios M, Shaw JG. Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect Immun. 2004;72:1939-45.
- 21. Leal NC, Sobreira M, Leal-Balbino TC, Almeida AM, Silva MJ, Mello DM, et al Evaluation of a RAPD-based typing scheme in a molecular epidemiology study of Vibrio cholerae O1, Brazil. J Appl Microbiol. 2004;96:447-54.
- 22. Merino S, Shaw JG, Tomás JM. Bacterial lateral flagella: an inducible flagella system. FEMS Microbiol Lett. 2006;263:127-35.
- 23. Nawaz M, Khan SA, Khan AA, Sung K, Tran Q, Kerdahi K, et al Detection and characterization of virulence genes and integrons in Aeromonas veronii isolated from catfish. Food Microbiol. 2010;27:327-31.
- 24. Nelson EJ, Harris JB, Morris JG Jr, Calderwood SB, Camilli A. Cholera transmission: the host, pathogen and bacteriophage dynamic. Nat Rev Microbiol. 2009;10:693-702.
- 25. Noguerola I, Blanch AR. Identification of Vibrio spp. with a set of dichotomous keys. J Appl Microbiol. 2008;105:175-85.
- 26. Pemberton JM, Kidd SP, Schmidt R. Secreted enzymes of Aeromonas FEMS Microbiol Lett. 1997;152:1-10.
- 27. Sen K, Rodgers M. Distribution of six virulence factors in Aeromonas species isolated from US drinking water utilities: a PCR identification. J Appl Microbiol. 2004;97:1077-86.
- 28. Soler L, Figueras MJ, Chacón MR, Vila J, Marco F, Martinez-Murcia AJ, et al Potential virulence and antimicrobial susceptibility of Aeromonas popoffii recovered from freshwater and seawater. FEMS Immunol Med Microbiol. 2002;32:243-7.
- 29. Vilches S, Jiménez N, Merino S, Tomás JM. The Aeromonas dsbA mutation decreased their virulence by triggering type III secretion system but not flagella production. Microb Pathog. 2012;52:130-9.
- 30. World Health Organization. Guidelines for cholera control. Geneva: WHO; 1993. ISBN: 92 4 154449 X.
Publication Dates
-
Publication in this collection
12 Nov 2012 -
Date of issue
Dec 2012
History
-
Received
09 Jan 2012 -
Accepted
05 June 2012