Abstracts
To obtain the recombinant group 2 allergen product of Dermatophagoides farinae (Der f 2), the Der f 2 gene was synthesized by RT-PCR. The full-length cDNA comprised 441 nucleotides and was 99.3% identical to the reference sequence (GenBank AB195580). The cDNA was bound to vector pET28a to construct plasmid pET28a(+)-Der f 2, which was transformed into E. coli BL21 and induced by IPTG. SDS-PAGE showed a specific band of about 14kDa in the hole cell lysate. s estiated by chroatography, about 3.86 g of the recobinant product as obtained, which conjugated with serum IgE from asthmatic children. The protein had a signal peptide of 17 amino acids. Its secondary structure comprised an alpha helix (19.86%), an extended strand (30.82%), and a random coil (49.32%). The subcellular localization of this allergen was predicted to be at mitochondria. Furthermore, its function was shown to be associated with an MD-2-related lipid-recognition (ML) domain. The results of this study provide a solid foundation for large-scale production of the allergen for clinical diagnosis and treatent of allergic disorders.
Dermatophagoides farinae; house-dust-mite allergy; mite allergens; Der f 2; allergen engineering; immunotherapy; bioinformatics
Com a finalidade de obter o produto recombinante do alergeno grupo 2 do Dermatophagoides farinae (Der f2), o gene Der f2 foi sintetizado por RT-PCR. O cDNA continha 441 nucleotídeos e era idêntico em 99,3% à sequência de referência (GenBank AB195580). O cDNA foi ligado ao vetor pET28a para construir o plasmídeo pET28a(+)-Der f2, o qual foi introduzido por transformação em E. coli BL21 e induzido por IPTG. Em SDS-PAGE foi vista mia banda específica de 14 kDa no lisado celular. Conforme estimado por cromatografia, cerca de 3,86 mg do produto recombinante foi obtido, que reagia com IgE sérica de crianças asmáticas. A proteína continha um peptídeo sinal de 17 amino ácidos. Sua estrutura secundária consistia de uma alfa hélice (19,86%), uma fita estendida (30,82%), e uma sequência randômica (49,32%). A localização subcelular desse alergeno foi predita ocorrer nas mitocôndrias. Sua função foi associada com o domínio de reconhecimento lipídico (ML) relacionado a MD-2. Os resultados desse estudo permitem a produção em larga escala do alergeno para o diagnóstico clínico e tratamento das doenças alérgicas.
Dermatophagoides farinae; alergia a ácaro de poeira doméstica; alergenos acarianos; Der f 2; alergeno recombinante; imunoterapia; bioinformática
BIOMEDICAL AND MEDICAL SCIENCES
Cloning, expression, and analysis of the group 2 allergen from Dermatophagoides farinae from China
Yu-bao Cui; Ying Zhou; Weihong Shi; Guifang Ma; Li Yang; Yungang Wang
Department of Laboratory Medicine, Yancheng Health Vocational & Technical College, Jiangsu Yancheng 224006, P.R. China
Correspondence to Correspondence to: Yu-bao Cui E-mail: ybcui1975@hotmail.com
ABSTRACT
To obtain the recombinant group 2 allergen product of Dermatophagoides farinae (Der f 2), the Der f 2 gene was synthesized by RT-PCR. The full-length cDNA comprised 441 nucleotides and was 99.3% identical to the reference sequence (GenBank AB195580). The cDNA was bound to vector pET28a to construct plasmid pET28a(+)-Der f 2, which was transformed into E. coli BL21 and induced by IPTG. SDS-PAGE showed a specific band of about 14kDa in the hole cell lysate. s estiated by chroatography, about 3.86 g of the recobinant product as obtained, which conjugated with serum IgE from asthmatic children. The protein had a signal peptide of 17 amino acids. Its secondary structure comprised an alpha helix (19.86%), an extended strand (30.82%), and a random coil (49.32%). The subcellular localization of this allergen was predicted to be at mitochondria. Furthermore, its function was shown to be associated with an MD-2-related lipid-recognition (ML) domain. The results of this study provide a solid foundation for large-scale production of the allergen for clinical diagnosis and treatent of allergic disorders.
Key words:Dermatophagoides farinae, house-dust-mite allergy, mite allergens, Der f 2, allergen engineering, immunotherapy, bioinformatics.
RESUMO
Com a finalidade de obter o produto recombinante do alergeno grupo 2 do Dermatophagoides farinae (Der f2), o gene Der f2 foi sintetizado por RT-PCR. O cDNA continha 441 nucleotídeos e era idêntico em 99,3% à sequência de referência (GenBank AB195580). O cDNA foi ligado ao vetor pET28a para construir o plasmídeo pET28a(+)-Der f2, o qual foi introduzido por transformação em E. coli BL21 e induzido por IPTG. Em SDS-PAGE foi vista mia banda específica de 14 kDa no lisado celular. Conforme estimado por cromatografia, cerca de 3,86 mg do produto recombinante foi obtido, que reagia com IgE sérica de crianças asmáticas. A proteína continha um peptídeo sinal de 17 amino ácidos. Sua estrutura secundária consistia de uma alfa hélice (19,86%), uma fita estendida (30,82%), e uma sequência randômica (49,32%). A localização subcelular desse alergeno foi predita ocorrer nas mitocôndrias. Sua função foi associada com o domínio de reconhecimento lipídico (ML) relacionado a MD-2. Os resultados desse estudo permitem a produção em larga escala do alergeno para o diagnóstico clínico e tratamento das doenças alérgicas.
Palavras-chave:Dermatophagoides farinae, alergia a ácaro de poeira doméstica, alergenos acarianos, Der f 2, alergeno recombinante, imunoterapia, bioinformática.
INTRODUCTION
Dust mites have been recognized to be associated with the rising incidence of asthma and other allergic diseases (Milián and Díaz 2004, Nadchatram 2005). At present, mite extract is used in clinical diagnosis and treatment of allergic diseases, especially in developing countries. However, the extract is a complex mixture of allergens, non-allergic or toxic proteins, and some enzymes, which have been reported to be associated with flushing, swelling, induration, necrosis and other local reaction, shock, laryngeal edema, bronchospasm, urticaria, angioedema, generalized erythema, and other systemic side effects (Mauro etal. 2006, Zhang et al. 2009). Over the past 10 years, some groups of mite allergens have been cloned, sequenced, expressed and purified. The three-dimensional structures of some allergens were determined, and B and T-cell epitopes were mapped (Jeong et al. 2006, Mothes et al. 2006). The recombinant allergens were produced in bacteria, yeasts and insect cell expression systems, and have been demonstrated to have good IgE binding activity similar to their natural counterparts. They have also shown good effects in skin prick tests and other allergen diagnosis methods (Jeong et al. 2006, Mothes et al. 2006). These recombinant allergens are intended to replace natural allergens and to allow the development of new immunotherapy projects, including the use of hypoallergens, allergens coupled to IgE suppressive adjuvants, and peptide-based therapies (Dinakar and Portnoy 2004, Linhart and Valenta 2005).
Polymorphisms have been reported in dust mite allergens. Therefore, recombinant allergens should be produced according to thorough study of the prevalent allergens in different regions (Linhart and alenta 2005). In our previous study, we obtained the gene encoding the group 2 allergen from Dermatophagoides farinae (Der f 2). An additional region of 87 bp (from 77 to 163 bp) was present in our strain that was absent from the reference sequence (GenBank AB195580) (Cui et al. 2007). We attempted to express this gene in Escherichia coli (E. coli) with no success. We, then, isolated total RNA from Dermatophagoides farinae again, and synthesized the cDNA encoding Der f 2 by RTPCR. We obtained a Der f 2 gene that was identical in length to the reference sequence, and successfully expressed it in E. coli.
MATERIALS AND METHODS
PATIENT SERA
Five asthmatic children were involved in this study. They were demonstrated to be allergic to dust mites by the skin prick test with Dermatophagoides farinae extract provided by the Department of Parasitology, Medical College of Fudan University. Blood (5 ml) was extracted from each child and centrifuged for serum. We mixed five sera samples and treated the mixture as the positive control in western-blotting. On the other hand, umbilical serum samples were taken from five children whose mothers had no atopic history, centrifuged, mixed, and used as negative controls.
PREPARATION OF CDNA AND POLYMERASE CHAIN REACTION (PCR)
According to our previous report (Cui et al. 2007, 2008), house dust mites were cultured and isolated. Total RNA was obtained using RNA isolator (TaKaRa Biotech Co. Led, Dalian, China, Code No. D312) and stored at -80°C. For PCR amplification of the cDNA encoding Der f 2, two primers were designed and synthesized based on the GenBank sequence AB195580. The forward primer sequence was (5'GGATCCATGATTTCCAAAATCTTGTGCC3'), and the reverse was (5'CTCGAGTTAATCACGGATTTTACCATGG3'). These two primers had a BamH I site and a Xho I site at their 5' ends (underlined), respectively. Reverse transcription (RT) was performed using the total RNA isolated from mites with High Fidelity PrimeScript™ RT-PCR Kit (TaKaRa Biotech Co. Led, Code No. DR027A) in the PCR Thermal Cycler Dice (TaKaRa Biotech Co. Led, Code No. TP 600). The reaction mixture for RT had total RNA (2 μl), 20 mM of the reverse primer (1 μl), 10 mM of dNTP Mixture (1 μl), 20 mM of Random 6-mers (1 μl), and RNase Free H2O (5 μl), which was set at 65 °C for 5 min, followed by ice-bath for 2 min. Then, 5 x PrimeScript RT Buffer (4 ml), 40 U/mL of RNase Inhibitor (0.5 ml), PrimeScript RTase (0.5 μl), and RNase Free dH2O (5 μl) were added in. 20 ml of the final reaction mixture was incubated at 30°C for 10 min, 42°C for 30 min and 95°C for 5 min. The RT product was used as a template for PCR in the same thermal Cycler Dice with PrimeSTAR® HS DNA Polymerase (TaKaRa Biotech Co. Led, Code No. DR010A). The total reaction mixture had RT products (2 μl), 5x PrimeSTAR PCR Buffer (10 μl), 2.5 mM of dNTP Mixture (4 μl), 20 mM of the forward primer (1 μl), 20 mM of the reverse primer (1 μl), 2.5 U/μL of PrimeSTAR HS DNA Polymerase (0.5 μl), and dH2O (31.5 ml). PCR conditions used here included an initial incubation for 3 min at 94°C, followed by 30 cycles of 10 sec at 98°C, 15 sec at 55°C, and 30 sec at 72°C. After a final incubation for 10 min at 72°C, the amplicons were analyzed by agarose gel electrophoresis (1.0%) and visualized with ImageMaster® VDS.
CLONING AND DNA SEQUENCING
The PCR-amplified DNA was recovered from the gel with Agarose Gel DNA Purification Kit Ver. 2.0 (TaKaRa Biotech Co. Led, Code No. DV805). A poly-A tail was added with DNA A-Tailing Kit (TaKaRa Biotech Co. Led, Code No. D404), and the products were bound to pMD19-T simple vector (TaKaRa Biotech Co. Led, Code No. D104). E. coli JM109 (TaKaRa Biotech Co. Led, Code No. D9052) was, then, transformed with the recombinant plasmids; positive clones were selected by blue/white screening on Luria-Bertani (LB) plates containing 100 mg/ml ampicillin, and confirmed by restriction enzyme analysis and automatic DNA sequencing.
CONSTRUCTION OF EXPRESSION PLASMIDS pET28a (+)-Derf2
The recombinant plasmids constructed above were digested with BamH I and Xho I to release the cDNA fragment encoding Der f 2. The enzyme digestion reaction mixture had pMD19-T-Der f 2 10 ml, 10 U/ml of BamH I (5 ml), 10 U/ml Xho I (5 ml), 10x K Buffer (10 ml), and dH2O (70 ml), which was set at water-bath for four hours at 37°C. The digested fragment was subjected to agarose gel electrophoresis and recovered from the gel using Agarose Gel DNA Purification Kit Ver. 2.0 (TaKaRa Biotech Co. Led, Code No. DV805). The fragment was, then, subcloned into expression vector pET28a(+) (Novagen, Germany, Kit Lot No. N72770) to create pET28a(+)-Der f 2 using the DNA Ligation Kit (TaKaRa Biotech Co. Led, Code No. D6023). E. coli Competent Cells JM109 (TaKaRa Biotech Co. Led, Code No. D9052) were transformed with pET28a(+)-Der f 2 plasmids; positive clones were selected by blue/ white screening and verified by restriction enzyme analysis with BamH I and Xho I.
EXPRESSION of RDER F 2 in E. coli BL21 (DE3)
5 μl of pET28a (+)-Der f 2 plasmid were purified using the MiniBEST Plasmid Purification Kit Ver. 2.0 (TaKaRa Biotech Co. Led, Code No. DV801A) and transformed into 100 ml of E. coli BL21 (DE3, Stratagene, USA). E. coli BL21 carrying pET28a (+)-Der f 2 was grown on LB plates containing 50 mg/ml of kanamy-cin at 37°C overnight. A single colony was inoculated into 2 ml LB plates at 37°C overnight, 100 ml of which was taken and added into a glass tube containing 5 ml LB fluid containing kanamycin and, then, cultured at 37°C. 100 mM of isopropyl-yβ-D-thiogalactopyranoside (IPTG, 50 μl, final 1 mM) was added to induce the lac promoter. The E. coli cells were harvested by centrifugation, and PBS Buffer (200 μl/tube) was added in. After centrifugation and resuspension, ultrasonic disruption was used until the obtention of a transparent fluid, 50 μl of which was taken as the whole cell lysate. The lysate was centrifuged and the supernatant and pellet were regarded as the soluble and insoluble protein samples, respectively. The whole cell lysate (10 μl), soluble and insoluble protein samples were taken individually, and 2.5 μl of 5x SDS sample buffer were added into each of them. The samples were heated for 10 min at 95°C and subjected to sodium dodecylsulfate-polyacryl-amide gel electrophoresis (SDS-PAGE) with 12.5% of polyacrylamide gel and CBB-R250 staining.
ISOLATION, PURIFICATION, AND RENATURATION OF RECOMBINANT RDER F 2 PROTEINS
E. coli BL21 carrying pET28a (+)-Der f 2 was grown on LB plates containing 50 mg/ml of kanamycin at 37°C overnight. A single colony inoculated into 5 ml of LB fluid with shaking at 250 rpm overnight at 37°C was treated as the seed fluid for fermentation. 50 ml of the seed fluid were inoculated into 2000 ml LB fluid containing 50 μg/ml of kanamycin at 30°C shaking at 250 rpm for fermentation. 100 mM IPTG were added until the final concentration reached 1 mmol/l. The induced cells were collected, suspended in PBS (pH 7.4), and broken by ultrasonic waves in an ice bath until the fluid became transparent. This fluid with broken cells was centrifuged at 7000 rpm at 4°C for 15 min to collect rough inclusion bodies, which were then washed in PBS buffer containing 2 mol/l of urea for the purified product. The purified inclusion bodies were resuspended in PBS buffer (pH 8.0) containing 8 mol/L of urea and 5 mmol/l of DTT, stirred slightly at 4°C overnight, and centrifuged at 1000 rpm for 10 min. The supernatant was filtered by microfiltration membrane with a pore size of 0.22 μm. This protein product was further purified by nickel-affinity chromatography at a speed of 4.0 ml/min, eluted with 25, 50, 75, 100, 125, and 150mmol/Lof imidazole solution at a speed of 6 ml/min. The elution peak at each stage was collected and determined for molecular weight and purity by SDS-PAGE. All of the elution fractions were freeze-dried for protein power. Lastly, the product was re-natured by gradient dialysis with a renat-uration solution containing 2 mol/l to 0.2 mol/L of urea, and 0.2 mol/l of EDTA continuously.
WESTERN BLOTTING
For western blotting analysis, proteins were transferred to a PVDF membrane (TIANGEN, Biotech Co. Led, Beijing, China) and incubated with primary antibodies kept in the sera from asthmatic children allergic to dust mites at 4°C overnight. They were, then, incubated with a secondary antibody (Horse anti-human IgE, AP conjugated, ZYMED Laboratories, USA) for 1 hr at room temperature, and the proteins were visualized using 3 ml of BCIP/NBT (Roche, Switzerland).
NUCLEOTIDE SEQUENCE, INFERRED AMINO ACID SEQUENCE, STRUCTURAL AND FUNCTION ANALYSIS, ALIGNMENT AND PHYLOGENY
Sequences were edited to remove the vector sequence and the extra restriction sites. The open reading fragment (ORF) was obtained using the ORF finder in the NCBI (National Center for Biotechnology Information) website. The amino acid sequence of Der f 2 was deduced using Translate Tools, and its physiochemical properties were predicted by ProtParam Tools in ExPaSy web server. The signal peptide of Der f 2 was analyzed by the SignalP 3.0 server. The functional site was searched for in the InterPro database in EBI (European Bioin-formatics Institute) website. The subcellular localization for Der f 2 was predicted by CELLO v.2.5, and its secondary structure was analyzed by GOR4.0 software. The published sequences that matched our sequenced cDNA were obtained from GenBank using BLAST. The published sequences of ticks and mites were chosen for alignment using ClustalW2 at the EBI website, and computed for similarity by VECTOR NTI 9.0 software (IBI, New Haven, CT, USA). A phylogenetic tree was constructed with maximum parsimony methods in Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0 software.
RESULTS
CLONING AND SEQUENCING OF cDNA ENCODING DER f 2
Total RNA was isolated from adult mites, and Der f 2 cDNA fragments were amplified by RT-PCR, obtaining a 441 bp product (Fig. 1A). The recovered PCR product was cloned into vector pMD19-T and transformed into E. coli competent cells (JM109) and positive clones. The product was confirmed by restriction enzyme digestion and automatic DNA sequencing (Fig. 1B, the vector sequence and the added restriction sites were removed from the sequencing results). The sequencing results showed that our Der f 2 cDNA is 99.3% identical to the previously published sequence (GenBank AB195580). There is one mutation at position 328 (G>A) resulting in amino acid mutation from Aspartate to Asparagine, and the other two mutations at positions 382 (G>A) and 384 (T> C) resulting in amino acid mutation from Valine to Isoleucine. Using the ORF Finder, a completeORF was found within the Der f 2 cDNA; its length is 441 bp from the start codon ATG to the stop codon TGA (Fig. 1B).
EXPRESSION AND PURIFICATION OF RECOMBINANT PROTEIN DER f 2
The cDNA encoding Der f 2 allergen was excised from recombinant plasmid pMD19-T-Der f 2, sub-cloned into expression vector of pET28a (+) and confirmed by restriction analysis (). E. coli BL21 was transformed with plasmid pET28a (+)-Der f 2, and protein expression was induced with IPTG. We observed a single band of about 14 kDa on SDS-PAGE, which agreed with the predicted molecular weight of 14.076 kDa for the Der f 2 protein (Fig. 2B). E. coli BL21 carrying pET28a (+)-Der f 2 was recovered from a 2000 ml of fermentation solution, collected, ultrasonically disrupted, and centrifuged. The precipitate was washed, dissolved and filtered. The supernatant was purified by nickel-affinity chromatography and eluted with imidazole solution, and the pure protein presented in 25, 50, 75, 100, and 125 mmol/l of elution fluid was shown by SDS-PAGE (Fig. 2C). These elution fractions were collected and freeze-dried, and 3.86 mg of protein power was obtained. The recombinant protein can bind the IgE antibody in asthmatic children by western-blotting (Fig. 2D), which demonstrated that the expression product has good allergenicity.
BIOINFORMATICS FOR AMINO ACID COMPOSITION, PHYSIOCHEMICAL PROPERTIES, SECONDARY STRUCTURE, SUBCELLULAR LOCALIZATION, AND FUNCTIONAL SITE OF THE RECOMBINANT PROTEIN rDER f 2
To identify the physiochemical properties of the recombinant protein, the sequenced result was translated into an amino acid sequence, which should encode a protein comprising 146 amino acids with a 17 amino acid signal peptide analyzed by SignalP 3.0 software. After the removal ofthe signal peptide sequence, the mature allergen comprised 129 amino acids residues with a relative molecular weight of 14.076, an isoelectric point of 6.46, a molar extinction coefficient of 8970, and an instability index of 21.31. The complete pro-protein was computed for a Grand average of hydropathicity (GRAVY) of 0.105 by ProtParam Tools, which indicated that it should be hydrophilic. With regard to Der f 2 secondary structure, GOR4 predicted that 19.86% (29 aa) of the protein were in alpha helices, 30.82% (45 aa) in extended strands, and 49.32% (72 aa) in random coils (Fig. 3). Using CELLO v.2.5, the subcellular localization of Der f 2 from China was concluded to be mitochondrial (Table I). A MD-2-related lipid-recognition (ML) domain from residue 22 to 141 was found in the Der f 2 pro-protein by InterProScan.
AMINO ACID SEQUENCE HOMOLOGY ANALYSIS, ALIGNMENT, AND PHYLOGENY
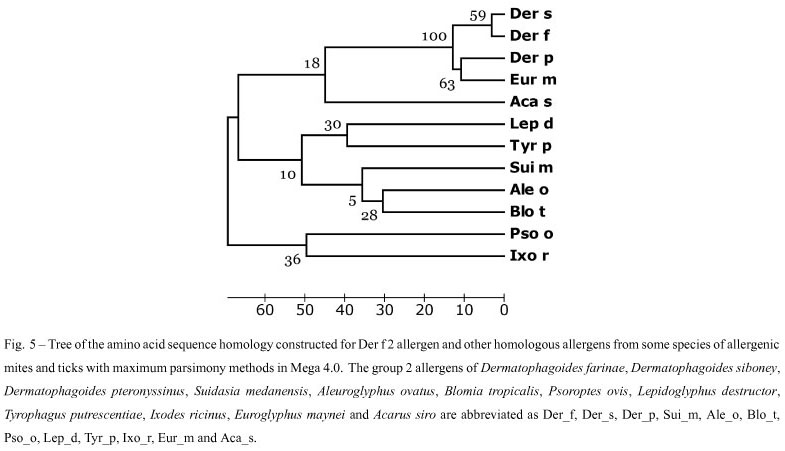
The homology between the deduced amino acid sequence of Der f 2 in this paper and other proteins was determined by comparing the sequence with all non-redundant GenBank CDS translations + PDB + Swiss-Prot + PIR + PRF (excluding environmental samples) using BLASTp at the NCBI website. Based on our BLASTp search results, similar amino sequences form ticks and mites, i.e. Dermatophagoides siboney (GenBank ABC96702), Dermatophagoides pteronyssinus (GenBank CAK22338), Suidasia medanensis (GenBank AAS75831), Aleuroglyphus ovatus (GenBank ABU97461), Blomia tropicalis (GenBank ABG76185), Psoroptes ovis (GenBank Q965E2), Lepidoglyphus destructor (GenBank P80384), Tyrophagus putrescentiae (GenBank ABU97478), Ixodes ricinus (GenBank ABL61-513), Euroglyphus maynei (GenBank Q9TZZ2) and Acarus siro (GenBank ABU97459) were chosen. After sequences coding for signal peptides were deleted, they were aligned using the ClustalW2 (Fig. 4), and a phylogenetic tree was constructed using Mega 4.0 software. The similarity between Der f 2 and Der p 2, Der s 2 and Eur m 2 were 88%, 96% and 83% (Table II), and these four mites were clustered with 100% of Bootstrap values (Fig. 5).
DISCUSSION
More than 30 groups of allergens were found in mite extracts, including enzymes, enzyme inhibitors, and structural proteins, which were named according to the order in which they were discovered. Group 2 allergens are regarded as a major cause of mite allergy (Thomas et al. 2002, 2004). In this study, we obtained a Der f 2 cDNA fragment with 99.3% identity with the reference sequence, by conventional RT-PCR. When the plasmid pET28a (+)-Der f 2 was transfomied into E. coli BL21, the obvious bands of about 14 kDa were observed in the whole cell lysate and the pellet of cells on SDS-PAGE, which agreed with the predicted result and suggest that the prokaryotic expression system for Der f 2 was successfully developed, as well as the product presented in the inclusion bodies. The expression vector PET28a(+), with a total length of 5369bp, expressed the recombinant protein containing the six-histidine tail, which bound to the nickel resin and could be eluted with a chelating agent after other proteins were removed with the appropriate buffers. The single specific reaction band of about 14 kDa was present in the 25, 50, 75, 100, and 125 mmol/l of imidazole elution fractions on SDS-PAGE. Furthermore, the purified recombinant allergen could bind to IgE in sera from asthmatic children, which showed that the recombinant protein had good allergenicity and was a good foundation for the production of genetic allergens.
The cDNA sequence for Der f 2 was translated into an amino acid sequence, and the signal peptide sequence was deduced at position 1 to 17. After the removal of the signal peptide, the mature protein was predicted to be a hydrophilic protein of 129 residues and 14.076 kDa of molecular weight. Interestingly, the subcellular location for this protein was predicted to be mitochondrial according to its N-peptide, physicochemical and neighboring sequence compositions, by CELLO2.5. Additionally, the group 2 allergen was predicted to have a MD-2-related lipid-recognition (ML) domain, which is associated with lipid recognition, particularly with the recognition of pathogen-related products. The ML domain has an immunoglobulin-like beta-sandwich fold similar to that of E-set Ig domains.
Many studies have been published concerning house dust mites, which belong to the family Pyroglyphi-dae, especially Dermatophagoides pteronyssinus, Dermatophagoides farinae, and Euroglyphus maynei. Other mite species, referred to as "storage mites" because of their consumption of stored food, are also considered allergenic, although their study is more limited (Fernández-Caldas et al. 2007). We used the deduced amino acid sequence of Der f 2 from China to search the protein databases at NCBI using BLASTp. Similar amino acid sequences of other mite species were identified, including D. siboney, D. pteronyssinus and Euroglyphus maynei listed in Pyroglyphidae; Lepidoglyphus destructor and Blomia tropicalis in Glycyphagidae; Suidasia medanensis, Aleuroglyphus ovatus, Tyrophagus putrescentiae and Acarus siro in Acaridae. Besides Dermatophagoides spp. other mite-species, especially Blomia tropicalis due to its abundance in tropical and subtropical regions, were previously considered to be important from economic and sanitary perspectives, but are now being recognized as important contributors to the allergen content in house dust. They can cause occupational respiratory allergies in farmers and other occupa-tionally exposed individuals (Nadchatram 2005). Many of these allergens have shown sequence homology and biological functional similarity to those described in Dermatophagoides spp. The amino acid sequence similarity between the group 2 allergen of D. farinae and that of D. pteronyssinus, D. siboney and E. maynei were calculated at 88%, 96%, and 83%, respectively, in this paper. In the phylogenetic tree constructed by Mega 4.0, D. farinae, D. pteronyssinus, D. siboney and E. maynei were clustered with 100% of bootstrap values, which agreed with the present taxonomy. D. farinae and D. siboney were clustered with 59% of bootstrap values, and D. pteronyssinus and Euroglyphus maynei with 63%. Generally, if the bootstrap value is below 70%, the relationship in the phylogenetic tree will be out of accord with the real taxonomy. In our previous report (Cui et al. 2008), Der p1 shared more than 87% identity in an amino acid sequence with Eur m1, but only 80% with Der f 1. D. pteronyssinus as evolutionarily closer to E. maynei than to D. farinae, even though D. pteronyssinus and D. farinae belong to the same genus (Dermatophagoides) in the phylogenetic tree constructed for the group 1 allergens from different mite-species. Therefore, the relationship among the common mite-species should be investigated using much more molecular data.
ACKNOWLEDGMENTS
This work was supported by National Sciences Foundation of China (NSFC 30060166), and Health Department of Jiangsu Province in China (Grant Number: Z200914 and J200907)
Manuscript received on March 3, 2010; accepted for publication on July 6, 2010
Present address: Department of Physics, University of Texas at San Antonio, One UTSA Circle, San Antonio, Texas 78249 USA.
- CUI YB, PENG JL, ZHOU P, PENG M and QIAN SY. 2007. Bioinformatic studies on the group 2 allergens of Dermatophagoides farinae from China. Asian Pac J Allergy Immunol 25: 199-206.
- CUI YB, ZHOU P, PENG JL, PENG M, ZHOU Y AND LIN YZ. 2008. Cloning, sequence analysis, and expression of cDNA coding for the major house dust mite allergen, Der f 1, in Escherichia coli. Braz J Med Biol Res 41: 380-388.
- DINAKAR C AND PORTNOY JM. 2004. Allergen Immunotherapy in the prevention asthma. Curr Opin Allergy Clin Immunol 4: 131-136.
- FERNÁNDEZ-CALDAS E, IRAOLA V AND CARNÉS J. 2007. MOLECUlar and biochemical properties of storage mites (except Blomia species). Protein Peptide Lett 14: 954959.
- JEONG KY, HONGB CS AND YONG TS. 2006. Recombinant allergens for diagnosis and immunotherapy of allergic disorders, with emphasis on cockroach allergy. Curr Protein Pept Sci 7: 57-71.
- LINHART B AND VALENTA R. 2005. Molecular design of allergy vaccines. Curr Opin Immunol 17: 646-655.
- MAURO M, RUSSELLO M, ALESINA R, SILLANO V, ALESSANDRINI A, DAMA A, PASSALACQUA G AND SENNA G. 2006. Safety and pharmacoeconomics of a cluster administration of mite immunotherapy compared to the traditional one. Eur Ann Allergy Clin Immunol 38: 31-34.
- MILIÁN E AND DÍAZ AM. 2004. ALLERGy to house dust mites and asthma. P R Health Sci J 23: 47-57.
- MOTHES N, VALENTA R AND SPITZAUER S. 2006. Allergy testing: the role of recombinant allergens. Clin Chem Lab Med 44: 125-132.
- NADCHATRAM M. 2005. House dust mites, our intimate associates. Trop Biomed 22: 23-37.
- THOMAS WR, SMITH WA, HALES BJ, MILLS KL AND O'BRIEN RM. 2002. Characterization and immunobiology of house dust mite allergens. Int Arch Allergy Immunol 129: 1-18.
- THOMAS WR, SMITH WA AND HALES BJ. 2004. The allergenic specificities of the house dust mite. Chang Gung Med J 27: 563-569.
- ZHANG L, WANG C, HAN D, WANG X, ZHAO Y AND LIU J. 2009. Comparative study of cluster and conventional immunotherapy schedules with Dermatophagoides pteronyssinus in the treatment of persistent allergic rhinitis. Int Arch Allergy Immunol 148: 161-169.
Publication Dates
-
Publication in this collection
28 Feb 2011 -
Date of issue
Dec 2010
History
-
Accepted
06 July 2010 -
Received
03 Mar 2010