Abstracts
Chlosyne lacinia saundersii is one of the most important pests of sunflower and it is the main target of insecticides applications. Larvae were collected in Londrina (PR), Santa Maria (RS), Dourados (MS), Ribeirão Preto (SP), Brasília (DF), Barreiras (BA), Uberaba (MG) and Vilhena (RO). Genomic DNA was extracted and amplified with ten-mer primers, which produced 101 loci. The size of the RAPD amplicons ranged from 180 to 2564 bp. Polymorphism among populations ranged from 31% to 67%, with the highest polymorphisms of 57% and 67% being detected in Uberaba and Vilhena populations, respectively. Populations with the highest similarity determined with Dice coefficient were from Ribeirão Preto and Barreiras, while insects from Londrina showed the highest similarity among them. Gene flow of C. lacinia saundersii 1.1 was lower than those previously observed for the noctuid Anticarsia gemmatalis Hübner, suggesting that C. lacinia saundersii populations are more isolated than the ones of this noctuid. Through the Analysis of Molecular Variance (AMOVA), RAPD variance was 33.64% among geographical populations and 66.36% within populations. These results suggest that populations of C. lacinia saundersii are genetically structured.
amova; gene flow; molecular marker; sunflower pest
Chlosyne lacinia saundersii é uma das mais importantes pragas da cultura do girassol e o principal alvo das aplicações de inseticidas. As larvas foram coletadas em Londrina (PR), Santa Maria (RS), Dourados (MS), Ribeirão Preto (SP), Brasília (DF), Barreiras (BA), Uberaba (MG) e Vilhena (RO). O DNA genômico foi extraído e amplificado com dez primers, que produziram 101 locos. O tamanho das amplificações de RAPD variou de 180 a 2564 pb. O polimorfismo entre as populações variou de 31% a 67%, com maior polimorfismo 57% e 67%, detectado em populações de Uberaba e Vilhena, respectivamente. As populações com maior similaridade determinada com o coeficiente de Dice foram de Ribeirão Preto e Barreiras, enquanto os insetos coletados em Londrina apresentaram maior similaridade entre eles. O fluxo gênico de C. lacinia saundersii de 1,1 foi menor que o observado para a Anticarsia gemmatalis Hübner Noctuidae, sugerindo que as populações de C. lacinia saundersii estão mais isoladas do que estes noctuideos. Através da análise de variância molecular (AMOVA), RAPD a variação foi de 33,64% entre as populações geográficas e 66,36% dentro das populações. Estes resultados sugere que as populações de C. lacinia saundersii são geneticamente estruturadas.
amova; fluxo gênico; marcador molecular; praga do girassol
AGRARIAN SCIENCES
Genetic diversity of the sunflower caterpillar (Chlosyne lacinia saundersii Doubleday and Hewitson) (Lepidoptera: Nymphalidae) populations determined by molecular RAPD markers
Fabiane CunhaI; Daniel R. Sosa GómezII; Jose J. da SilvaII; Talita M. AlexandreI; Flávio MoscardiIII
IPós-Graduação em Entomologia, UFPR, Embrapa Soja, Laboratório de Patologia de Insetos Caixa Postal 231, 86001-970 Londrina, PR, Brasil
IIEmbrapa Soja, Laboratório de Patologia de Insetos, Caixa Postal 231, 86001-970 Londrina, PR, Brasil
IIIUniversidade Estadual de Londrina - Departamento de Agronomia Laboratório de Controle Microbiano de Insetos Caixa Postal 6001, 86051-990 Londrina, PR, Brasil
Correspondence to Correspondence to: Flávio Moscardi E-mail: fmoscardi@gmail.com
ABSTRACT
Chlosyne lacinia saundersii is one of the most important pests of sunflower and it is the main target of insecticides applications. Larvae were collected in Londrina (PR), Santa Maria (RS), Dourados (MS), Ribeirão Preto (SP), Brasília (DF), Barreiras (BA), Uberaba (MG) and Vilhena (RO). Genomic DNA was extracted and amplified with ten-mer primers, which produced 101 loci. The size of the RAPD amplicons ranged from 180 to 2564 bp. Polymorphism among populations ranged from 31% to 67%, with the highest polymorphisms of 57% and 67% being detected in Uberaba and Vilhena populations, respectively. Populations with the highest similarity determined with Dice coefficient were from Ribeirão Preto and Barreiras, while insects from Londrina showed the highest similarity among them. Gene flow of C. lacinia saundersii 1.1 was lower than those previously observed for the noctuid Anticarsia gemmatalis Hübner, suggesting that C. lacinia saundersii populations are more isolated than the ones of this noctuid. Through the Analysis of Molecular Variance (AMOVA), RAPD variance was 33.64% among geographical populations and 66.36% within populations. These results suggest that populations of C. lacinia saundersii are genetically structured.
Key words: amova, gene flow, molecular marker, sunflower pest.
RESUMO
Chlosyne lacinia saundersii é uma das mais importantes pragas da cultura do girassol e o principal alvo das aplicações de inseticidas. As larvas foram coletadas em Londrina (PR), Santa Maria (RS), Dourados (MS), Ribeirão Preto (SP), Brasília (DF), Barreiras (BA), Uberaba (MG) e Vilhena (RO). O DNA genômico foi extraído e amplificado com dez primers, que produziram 101 locos. O tamanho das amplificações de RAPD variou de 180 a 2564 pb. O polimorfismo entre as populações variou de 31% a 67%, com maior polimorfismo 57% e 67%, detectado em populações de Uberaba e Vilhena, respectivamente. As populações com maior similaridade determinada com o coeficiente de Dice foram de Ribeirão Preto e Barreiras, enquanto os insetos coletados em Londrina apresentaram maior similaridade entre eles. O fluxo gênico de C. lacinia saundersii de 1,1 foi menor que o observado para a Anticarsia gemmatalis Hübner Noctuidae, sugerindo que as populações de C. lacinia saundersii estão mais isoladas do que estes noctuideos. Através da análise de variância molecular (AMOVA), RAPD a variação foi de 33,64% entre as populações geográficas e 66,36% dentro das populações. Estes resultados sugere que as populações de C. lacinia saundersii são geneticamente estruturadas.
Palavras-chave: amova, fluxo gênico, marcador molecular, praga do girassol.
INTRODUCTION
The sunflower caterpillar Chlosyne lacinia saundersii Doubleday and Hewitson (1849) (Lepidoptera: Nymphalidae), a Neotropical species, is the main sunflower defoliator in Brazil (Ungaro 1981, Nakano et al. 1981, Gallo et al. 2002). It was first cited in Brazil by Maranhão (1945), in Piracicaba, SP, being further found in other states, as Paraná (Silva et al. 1968, Moscardi 1983, G.L. Villas Boas et al., unpublished data) Rio de Janeiro (Silva et al. 1968), Mato Grosso do Sul (Boiça Jr. et al. 1984) and São Paulo (Silva et al. 1968, Boiça Jr. et al. 1984). Depending on the infestation level, larvae may cause up to 100% defoliation (Nakano et al. 1981). The literature on C. lacinia saundersii is scanty and most of the papers is related to controlling this insect with chemical insecticides, with very few studies on its biology and ecology and none on the genetic structure of populations of this, insect. Papers related to genetic aspects are restricted to the genetics of color variation in larvae of C. lacinia saundersii (Gorodenski 1969), while Neck et al. (1971) reported the ocurrence of lethal genes associated to the orange-colored larvae. De Vries (1987) in Costa Rica reported that the larva of C. lacinia saundersii is polimorphic, with coloration ranging from black to orange. In Brazil, Gallo et al. (2002) described the larva of C. lacinia saundersii as of black coloration, while Zucchi et al. (1993) described the larva as orange in color and covered with black scoli and setae. As this insect is an agricultural pest and considering the sunflower expansion (from 77.000 ha to 94.000 ha. i.e., 22.1%) (Lazzaroto et al. 2005) in the last six years in Brazil, it has a favorable habitat in terms of food resources, which can make possible the increase in the genetic flow thus, reducing the genetic differentiation. Therefore, studies related to the population genetics would be important for the development of management strategies mainly in species that faced changes in their habitats (Lougheed et al. 2000).
In the present study we have chosen, among the available molecular techniques, to use the RAPD (Random Amplified Polymorphic DNA), as it identifies the DNA polymorphism amplified at random through the use of a sole initiator of arbitrary sequence without previous knowledge of the species genome moreover this technique is of reduced cost (Williams et al. 1990). Although this technique results in amplification of random sequences, several authors have demonstrated its efficiency to ditinguish genotypes in insect populations (eg. Carvalho and Vieira 2001, Oliveira et al. 2002, Lopes-da-Silva et al. 2004). Considering that C. lacinia saundersii is an insect of distinct geographical and climatic conditions, this study aimed to evaluate the intra and interspecific genetic variation in this insect, which can contribute to the development of proper strategies for the integrated management of this pest.
MATERIALS AND METHODS
INSECT COLLECTION
Second to fourth-instar C. lacinia saundersii larvae were collected in eight Brazilian states, (Table I). Collections were made from December 2007 to June 2008 in sunflowers or in other host plants and sent to the Laboratory of Entomology of Embrapa Soja, Londrina, PR, Brazil. Larvae were kept in environmental chambers (FANEM, model 347 CDG, São Paulo, SP) at 26°C ± 1°C. 65% ± 5% UR, and 12h of photophase), and monitored to record for the occurrence of parasitoids and entomopathogens. Last instar larvae were placed in 15 mL sealed plastic vials containing dehydrated silica gel, which were maintained at -15°C for further DNA extraction. Thirty larvae were used for each population, being 10 of the nigra, 10 of the bicolor, and 10 of the rufa coloration.
DNA EXTRACTION
The DNA was extracted according to the protocol of Rogers and Bendich (1988), with few modifications. The extraction was made from the larval head to avoid a possible contamination with parasitoids in the larval hemocele (D.R. Sosa Gómez, unpublished data). Each sample was macerated in 480μl of extraction buffer at a final concentration of 200mM Tris-HCl (pH 8.0), 70mM EDTA, 2M NaCl, and 1% β-Mercaptoethanol. After addition of 120μl of cetyltrimethyl ammonium bromide 10%, the samples were kept at 65°C for 5 min. Then, 6μl of Proteinase K (10 mg/ml) was added and the samples were incubated at 65 °C for 60 min. After that the samples were kept at environment temperature and centrifuged at 16.000 rpm for 15 min. The supernatant (500μl) was transferred to another microcentri-fuge tube and added to it the same volume of chloro-phorm/isoamyl alcool (24:1).
After homogenization, each sample was centrifuged at 16.000 rpm for 15 min and, then, the aqueous phase (400μl) was transferred to another tube where the nucleic acids were precipitated with the same volume of cold isopropanol plus 45% of the volume of ammonium acetate (10M). DNA samples were mixed by gently shaking the tube and kept at -20°C for 2 h at -4°C overnight. The samples were, then, centrifuged at 14.000 rpm for 15 min, discarding the supernatant and washing the pellets with 300μL of cold ethanol 70%. The pellets were dry, with the tubes inverted, in environment temperature for 10 min, being, then, re-suspended with 100.8μl of RNAse solution (40/μg/ml) plus TE. After that, the tubes were placed in an environmental chambers from 30 min to 2 h for RNA degradation. Aliquots of DNA were kept at -15°C and defrost when needed for DNA amplification.
The DNA was quantified by using a spectrophotometer Nanodrop® ND-1000 V3.5 (Wilmington, DE USA), and its integrity was evaluated through electrophoresis in agarosis 0.8% gel and colored with 10 mg/ml of ethidium bromide. The electrophoresis was conducted with buffer of 10mM of NaOH and pH adjusted to 8.5 with boric acid (Brody and Kern 2004). The DNA was visualized and the image was digitalized using a transluminator with a system of image capture L-PIX -ST and L-PIX IMAGE 7.1M Pixel. Images were captured with the software L-PIX IMAGE 1.0.1 (Loccus Biotecnologia, São Paulo - SP).
RAPD REACTIONS
The amplification reactions were conducted in a volume of 25μl containing about 9ηg of DNA, Milli Q sterile water, buffer 10×, 0.4μM of primer, 2.4 mM of MgCl2, 0.1mM of dNTP and Taq polymerase enzyme (Gibco BRE) (1U). The independent control of the reactions was conducted without the template DNA, when the amplification reaction with each primer was done. The primers that did not produce clear amplification were not considered in the analysis. The PCR was done using a thermocycler PTC 200 DNA EngineTM (MJ Research, Scientific Support, Hayward, CA) 3.8 version, with the following program: 45 cycles at 94°C for 15 sec, 39°C for 30 sec, and 72°C for 1 min, and a final extension of 72°C for 7 min. With a volume of 25μl, the product of RAPD was submitted to electrophoresis in agarose 1.3% and buffer SB 1× at 120 volts. The Lambda DNA, digested with the endonucleases EcoRI, HindIII and BamHI, was used as a marker of molecular weight. The gels were colored with 4.5μl of ethidium bromide at mg/ml, and the image digitalized by the system previously described.
DATA ANALYSIS
Digital photos from gels were visually analyzed depicting the presence (1) and absence (0) of amplified bands with the different primers, assuming the occurrence of two allels per locus. Only conspicuous and consistent bands, as well as their absence, were considered to generate the binary matrix. The similarity among samples was calculated using the Dice Coefficient by the formula 2h/(a+b), being "h" the number of observed bands and "a+b" the total number considering presence and absence of bands (Dice 1945). This matrix was submitted to cluster analysis using the unweighted pair-group method with aritmetic mean (UPGMA). The procedures were conducted through the program NTSYSpc 2.01 of Numerical Taxonomy (Rohlf 2000). To verify the fitness between the similarity matrix and the obtained dendrogram, the coefficient of cophenetic (r) was calculated according to Sokal and Rohlf (1962). The statistical stability of the groupings was estimated by the bootstrap analysis, with 1.000 replications, using the computer program Applied Maths Bionumerics (1998).
The structure of the populations was determined by the analysis of variance for molecular data (AMOVA) by using the program Arlequin (Schneider et al. 2000).
RESULTS
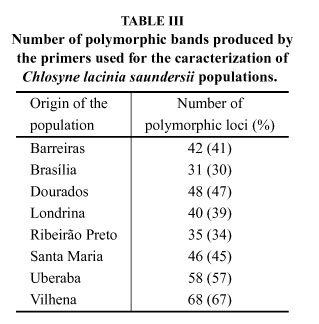
The ten used primers generated 101 loci, characterizing a high level of polymorphism in the C. lacinia saundersii populations. The number of obtained bands ranged from 5 (OPC-02) to 19 (OPA-05) (Table II, Fig. 1). Therefore, this last primer was the one that resulted in the highest polymorphism. The size of the RAPD products ranged from 180 to 2.564 pb. No monomorphic loci were observed among the studied populations despite being observed within each population (Table II)
The polyorphis aong the populations ranged from 31% to 67%, with the ones with the greater number of polymorphic loci (57% e 67%) being from Uberaba and Vilhena, respectively. However the smaller polymorphism was detected for the population from Brasília (31%), followed by the one from Ribeirão Preto (35%) (Table III).
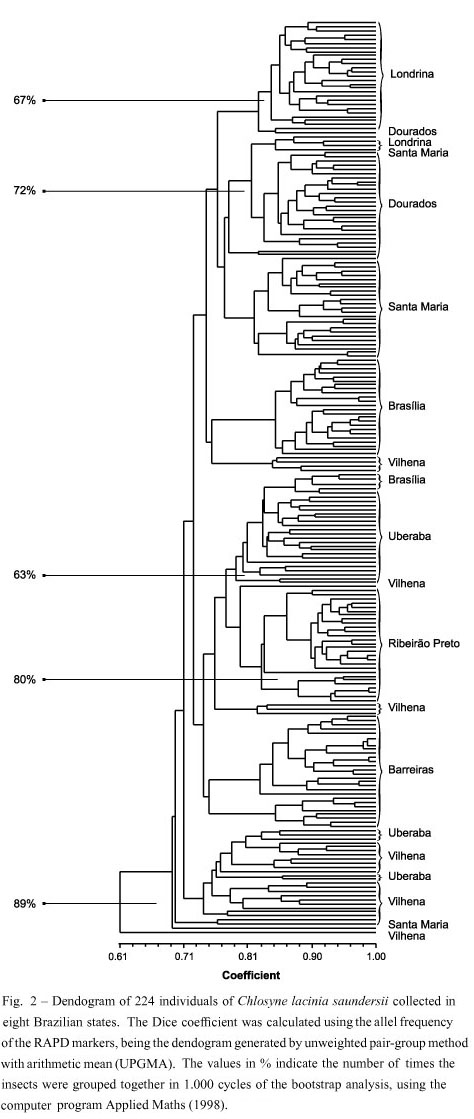
The largest proportion of individuals from Barreiras, Brasília, Ribeirão Preto, Dourados, Londrina, and Santa Maria formed six main groups according to their geographical location (Fig. 2). However, those from Uberaba and Vilhena presented similarities with individuals from other locations. The insects with the lowest similarity coefficient index were from the populations of Ribeirão Preto and Barreiras, while those with the highest siilarity ere collected in the region of Londrina (Fig. 2).
The populations with a close similarity among them were those from Londrina, Dourados and Santa Maria, which presented Dice coefficients ranging from 0.765 to 0.775 (Fig. 2). The most divergent population was the one from Vilhena, RO, with a Dice coefficient of 0.695. This population presented the highest value of polymorphic loci differently from the Brasília and Ribeirão Preto populations, which were the least polymorphic ones (Table III).
Larval color polyorphis as presented by orange larvae (rufa), those the ones with black as general color with orange dorsal spots (bicolor), and the ones totally black (nigra), as described by Edwards (1893). No differences on RAPD profiles were found among groups with the different color polymorphism pattern. The bootstrap analysis as consistent ith the Dice UPGMA dendogram for the clusters (Fig. 2). The AMOVA results presented a fixation index fst of 0.34 indicating that 33.64% of the genetic variation ocurred among populations, and 66.36% occurred within them (P<0.001) (Table IV).
DISCUSSION
Populations from Londrina, Dourados and Santa Maria were genetically close (Dice coef. = 0.76) compared to populations from Vilhena, Uberaba, Barreiras and Ribeirão Preto. The highest genetic dissimilarity (Dice coef. = 0.71-0.74) was observed between populations from Vilhena and Uberaba. The most heterogeneous population was from Vilhena, as samples from this population are found interspersed among populations from other localities.
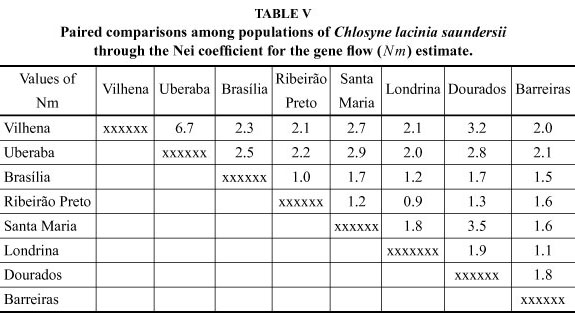
The gene flow indexes (Nm) ranged from 0.9 between the populations of Londrina and Ribeirão Preto, up to 6.7 between the populations of Vilhena and Uberaba (Table V). The gene flow index considering all the C. lacinia saundersii studied populations was 1.1, which is relatively low when compared, for instance, to populations of Anticarsia gemmatalis (Lepidoptera: Noctuidae), with a gene flow of 3.05 (Sosa-Gómez 2004). According to this author, the great flying capacity of this species would help to explain the high similarity among populations, since the geographical distances among them were large. Conversely, the similarity among populations of C. lacinia saundersii is lower and, consequently, there are lower interchange of genetic characteristics among them probably because the sunflower cultivated area is still small and patchy, which hampers the gene flow among populations.
The area cultivated with sunflower in Brazil is currently of 94.000 hectares, while soybean is grown over 21 million hectares (CONAB 2008). The continuity of soybean areas could be favoring a greater migration and, thus, a larger gene flow index of the velvetbean caterpillar (A. gemmatalis) in relation to the sunflower caterpillar due to the relative discontinuity of sunflower cultivated areas in the country. On the other hand, the sunflower caterpillar also feeds on alternative host plants, mainly of Asteraceae (Moscardi 1983, Campos-Farinha et al. 1997, Moscardi et al. 2005). However, there have been no studies related to the association of genotypes of C. lacinia saundersii with these natural plant hosts. On the other hand, there are known studies with the fall armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae), which confirmed a greater genetic similarity among populations obtained from a same plant host (McMichael and Pashley 1999, Busato et al. 2004). It suggests that the host plant represents an important factor in the natural selection process. Therefore, more detailed studies on geographical barriers, distribution, and abundance of these hosts can help to clarify the impact of host plants on C. lacinia saundersii genic flow.
Busato et al. (2004) evaluated the genetic diversity of S. frugiperda using AFLP markers and observed that studied populations were not different as there were overlapping of individuals from distinct populations in rice and/or corn. Almost nothing is known about the migration behavior of the sunflower caterpillar. Therefore, the origin and frequency of possible migrations are not known for this insect when occurring in sunflower and wild host plants in Brazil. C. lacinia saundersii occurs in diverse regions where sunflower is present in this country. Thus, it is possible that this species is well adapted to the different conditions of climate and topography.
Through the AMOVA, 34.64% of the total genetic variability could be attributed to differences among populations of C. lacinia saundersii, and 66.3% to differences within populations, which is an indicative that the studied populations were structured. Similar values were obtained with Sternechus subsignatus (Coleoptera: Curculionidae) that presented difference, among populations of 29% (Sosa-Gómez et al. 2008). For A. gemmatalis and S. frugiperda the variation among populations were of 14% and 18%, respectively (Sosa-Gómez 2004, Martinelli et al. 2006). These lower values may be related to the high gene flow in populations of these lepidopterous species.
The RAPD markers used in the present study were efficient for the genetic diferentiation of C. lacinia saundersii populations. This type of work is the only one done for this species up to the moment. However, additional studies with more specific molecular markers will be necessary to better elucidate how the species is distributed.
ACKNOWLEDGMENTS
The authors thank Dr. Vanoli Fronza for revising the manuscript, Dr. Eliseu Binneck for the statistical support, Silvana R. Rockenbach for the technical assistance; for insect collections, the following researchers: Débora Pires de Paula (Embrapa Recursos Genéticos e Biotecnologia), Mônica C. Pires and Pedro V. Lima (Fundação Bahia), Marino Alves (UNESP de Ribeirão Preto), Mar-ley Utumi (Embrapa Rondonia), Talita M. Alexandre and Jovenil J. da Silva (Embrapa Soja), Rejane C.R.K Roggia (UFSM), and Neylson Eustáquio Arantes (Embrapa Soja/Epamig - Uberaba); Adair V. Carneiro and Danilo Estevão for the support with the pictures and figures. This work was supported by Embrapa Soja and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Capes) and was approved for publication by the Publications Committee of Embrapa Soja with number 01/2009.
Manuscript received on September 17, 2009; accepted for publication on September 17, 2010
- APPLIED MATHS BIONUMERICS. 1998. The integral study of biological relationships. Manual, version 1.01, Kortrijk, Belgium, 154 p.
- BOIÇA JR AL, BOLONHEZI AC AND PACCINI NETO J. 1984. Levantamento de insetos pragas e seus inimigos naturais em girassol (Helianthus annuus L.), cultivadaem primeira e segunda época, no município de Sevíria-MS. An Soc Entomol Bras 13: 189-196.
- BRODY JR AND KERN SE. 2004. Sodium boric acid: a Tris-free, cooler conductive medium for DNA eletrophoresis. BioTechniques 36: 214-216.
- BUSATO GR, GRÜTZMACHER AD, OLIVEIRA AC, VIEIRA EA, ZIMMER PD, KOPP MM, BANDEIRA JM AND RODRIGUES TR. 2004. Análise da estrutura e diversidade molecular de populações de Spodoptera frugiperda (J.E. Smith, 1797) (Lepidoptera: Noctuidae) associadas ao milho e arroz no Rio Grande do Sul. Neotrop Entomol 33: 709-716.
- CAMPOS-FARINHA AEC, PINTO NPO AND GOVONE JS. 1997. Estudo do comportamento e desenvolvimento de lagartas de Chlosyne lacinia saundersii, Doubleday and Hewitson (1849) (Lepidoptera: Nymphalidae), no ataque a uma planta de girassol (Helianthus annuus L.). Arq Inst Biol 64: 143-147.
- CARVALHO AOR AND VIEIRA LGE. 2001. Determinação das condições ótimas para análises de PCR-RAPD em Atta sexdens rubropilosa Forel (Hymenoptera: Formicidae). Neotrop Entolmol 30: 593-600.
- COMPANHIA NACIONAL DE ABASTECIMENTO, CONAB. 2008. Acompanhamento da safra de girassol 2007/2008. Disponível em <http://www.conab.gov.br/conabweb/download/moc/titulos/T51s2007-2008.pdf>. de Vries PJ. 1987. The butterflies of Costa Rica and their natural history Papilionidae, Pieridae, Nymphalidae. Princeton University Press, 327 p.
- DICE LR. 1945. Measures of the amount of ecologic association between species. Ecology 26: 297-302.
- EDWARDS WH. 1893. Notes on a polymorphic butterfly, Synchloe lacinia, Geyer (in Hub. Zutr.), with description of its peparatory stages. Can Ent 25: 286-291.
- GALLO D ET AL. 2002. Entomologia Agrícola, Piracicaba, FEALQ, 920 p.
- GORODENSKI SA. 1969. The genetics of three polymorphic larval color forms of Chlosyne lacinia (Lepidoptera, Nymphalidae). Genet Res 14: 332-336.
- LAZZAROTO JJ, ROESSING AR AND MELLO HC. 2005. o agronegócio do girassol no mundo e no Brasil. In: Girassol no Brasil (Ed), Londrina: Embrapa Soja, p. 15-42.
- LOPES-DA-SILVA M, TONET GEL AND VIEIRA LGE. 2004. Characterization and genetic relationships among brazilian biotypes of Schizaphis graminum (Rondani) (Hemiptera: Aphididae) using RAPD markers. Neotrop Entomol 33: 043-049.
- LOUGHEED SC, GIBBS HL, PRIOR KA AND WEATHER-HEAD PJ. 2000. A comparison of RAPD versus micro-satellite DNA markers in population studies of the Mas-sasauga rattlesnake. J Hered 91: 458-463.
- MARANHÃO ZC. 1945. Chlosyne lacinia saundersii, praga do girassol. Rev da gricultura, Piracicaba 20(1-2): 199.
- MARTINELLI S, BARATA RM, ZUCCHI MI, SILVA FILHO MC AND OMOTO C. 2006. Molecular variability of Spo-doptera frugiperda (Lepidoptera: Noctuidae) Populations Associated to Maize and Cotton Crops in Brazil. J Econ Entomol 99: 519-526.
- MCMICHAEL M AND PASHLEY DP. 1999. Differences in amplified fragment-length polymorphisms in fall armyworm (Lepidoptera: Noctuidae) host strains. Ann Entomol Soc Am 92: 175-181.
- MOSCARDI F. 1983. Plantas Hospedeiras da lagarta do girassol, Chlosyne lacinia saundersii, no Estado do Paraná. In: Empresa BRASILEIRA DE PESQUISA AGROPECUÁRIA. CENTRO Nacional de Pesquisa de Soja, Londrina, PR. Resultados de Pesquisa de Girassol Londrina, p. 25-26.
- MOSCARDI F, Sosa-Gómez DR and Corso IC. 2005. Invertebrados associados ao girassol e seu manejo. In: Girassol no Brasil (Ed), Londrina: Embrapa Soja, p. 471-500.
- NAKANO O, SILVEIRA NETO S AND ZUCCHI RA. 1981. Entomologia Econômica. Piracicaba, Livroceres, 314 p.
- NECK RW, BUSH L AND DRUMOND BA. 1971. Epistasis, associated lethals and brood effect on larval colour polymorphism of the patch butterfly Chlosyne lacinia Geyer. Heredity 26: 73-84.
- OLIVEIRA CM, FUNGARO MH, CAMARGO LEA AND LOPES JRS. 2002. Análise comparativa da estabilidade de Dalbulus maidis (Delong and Wolcott) (Hemiptera: Ci-cadellidae) sobre diferentes métodos de preservação para uso de RAPD-PCR. Neotrop Entomol 31: 225-231.
- ROGERS SO AND BENDICH AJ. 1988. Extraction of DNA from plant tissues. Plant Mol Biol 6: 1-10.
- ROIILF FJ. 2000. Ntsys-Pc Numerical taxonomy and multivariate analysis system version 2.1. Exeter Software, Setauket, NY.
- SCHNEIDER S, ROESSLI D AND EXCOFFIER L. 2000. Arlequin, version 2.0: A software for population genetic data analysis. Genetics and Biomtry Laboratory, University of Geneva, Switzerland.
- SILVA AGA, GONÇALVES CR, GALVÃO DM, GONÇALVES AJL, GOMES J, SILVA MN AND SIMONI L. 1968. QUARTO CAtálogo dos insetos que vivem nas plantas do Brasil; seus parasitos e predadores. Rio de Janeiro, Ministério da Agricultura/laboratório central de patologia vegetal, 622 p.
- SOKAL RR AND ROHHLF FJ. 1962. The comparison of den-dograms by objective methods. Taxon 11: 30-40.
- SOSA GÓMEZ DR. 2004. Intraspecific variation and population structure of the Velvetbean Caterpillar, Anticarsia gemmatalis Hübner, 1818 (Insecta: Lepidoptera: Noctuidae). Genet Mol Biol 27: 378-384. Sosa-Gómez DR, Coronel N, Binneck E, Zucchi MI and Rosado-Neto G. 2008. RAPD and mitochondrial DNA analysis of the soybean stalk weevil, Sternechus subsignatus (Coleoptera: Curculionidae). Bull Entomol Res 98: 475-481.
- UNGARO MRG. 1981. Recomendações técnicas para o cultivo do girassol. Correio Agrícola Bayer 2: 314-319.
- WILLIAMS JG, KUBELIK AR, LIVAK KJ, RAFALSKI LA AND TINGEY SV. 1990. DNA polymorphism amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18: 6531-6535.
- ZUCCHI RA, SILVEIRA NETO S AND NAKANO O. 1993. Guia de identificação de pragas agrícolas, Piracicaba, FEALQ, 139 p.
Publication Dates
-
Publication in this collection
28 Feb 2011 -
Date of issue
Dec 2010
History
-
Received
17 Sept 2009 -
Accepted
17 Sept 2010