Abstracts
The tambaqui, Colossoma macropomum, is the most popular fish species used for aquaculture in Brazil but there is no study comparing genetic variation among native and farmed populations of this species. In the present study, we analyzed DNA sequences of the mitochondrial DNA to evaluate the genetic diversity among two wild populations, a fry-producing breeding stock, and a sample of fish farm stocks, all from the region of Santarém, in the west of the Brazilian state of Pará. Similar levels of genetic diversity were found in all the samples and surprisingly the breeding stock showed expressive representation of the genetic diversity registered on wild populations. These results contrast considerably with those of the previous study of farmed stocks in the states of Amapá, Pará, Piauí, and Rondônia, which recorded only two haplotypes, indicating a long history of endogamy in the breeding stocks used to produce fry. The results of the two studies show two distinct scenarios of tambaqui farming in the Amazon basin, which must be better evaluated in order to guarantee the successful expansion of this activity in the region, and the rest of Brazil, given that the tambaqui and its hybrids are now farmed throughout the country.
Tambaqui; mitochondrial DNA; genetic variability; Aquaculture
O tambaqui, Colossoma macropomum, é a espécie de peixes mais popularmente usada para a aquicultura no Brasil, mas não há nenhum estudo comparando a variação genética entre as populações nativas e de cultivo desta espécie. No presente estudo foram analisadas sequências de DNA mitocondrial para avaliar a diversidade genética entre duas populações selvagens, um plantel de produção de alevinos, e uma amostra de estoques de piscicultura, todos da região de Santarém, no oeste do estado do Pará. Níveis similares de diversidade genética foram encontrados em todas as amostras e, surpreendentemente, o plantel mostrou expressiva representação da diversidade genética registrada em populações selvagens. Estes resultados contrastam consideravelmente com os do estudo anterior de estoques cultivados nos estados do Amapá, Pará, Piauí, Rondônia, que registrou apenas dois haplótipos, indicando uma longa história de endogamia nas matrizes utilizadas para a produção de alevinos. Os resultados dos dois estudos mostram dois cenários distintos de aquicultura do tambaqui na Amazônia, que devem ser melhor avaliados, a fim de garantir o sucesso da expansão da atividade na região, e no resto do Brasil, já que o tambaqui e seus híbridos agora são cultivados em todo o país.
Tambaqui; mtDNA; variabilidade genética; Aquicultura
INTRODUCTION
The natural range of Colossoma macropomum Cuvier, 1818, known locally as Tambaqui, includes the Amazon and Orinoco river basins (Araujo-Lima and Goulding 1998). This species is one of the most important fishery resources in the Amazon region, where it is a staple of the diet of traditional riverside populations. In recent years, increasing pressure on the wild stocks of this species has led to significant reductions in the catch volume and of the size of the individuals harvested (Batista and Petrere-Jr 2003, Isaac and Ruffino 2003). In Brazil, the Environment Ministry classified the species as overexploited or threatened by overexploitation (statute no. 5 of May 21, 2004).
In contrast with wild stocks, supplies of tambaqui from aquaculture are increasing, and the species is now the most important farmed fish in northern Brazil, where it corresponds to more than 70% of the production of the region's inland aquaculture operations (Ostrensky et al. 2008). The farming of Tambaqui is an extremely important alternative to the exploitation of natural stocks. However, it requires careful planning in order to guarantee that, it is not only viable, but also has minimal impact on the species' wild populations. A major potential problem is the interaction between farmed specimens and wild population through accidental escapes or intentional releases from fish farms, in particular if the farmed stock is composed by hybrids.
The Tambaqui has been targeted in artificial hybridization programs over the past few decades in Brazil. This process involves a female Tambaqui mated with males of two other Characiform species, Piaractus mesopotamicus (Pacu) and Piaractus brachypomus (Pirapitinga) producing the hybrids known as Tambacu and Tambatinga, respectively. This hybridization aims to combine the characteristics of different species to produce an economically more viable organism, which is, in particular, faster growing and more resistant to disease and low temperatures (Fontes et al. 1990, Senhorini et al. 1988). Tambacu in particular is farmed in most states in Brazil, contributing expressively with the amount of fish produced by continental aquaculture in the country (Ostrensky et al. 2008).
In regard the genetic characterization of species with aquaculture potential, there have been some advances in the understanding of the genetic variability of wild and captive populations of Colossoma macropomum. Based on DNA sequences of the mitochondrial Control Region, these studies have shown that the wild populations constitute a panmictic unit, with high haplotype diversity, but low nucleotide diversity, and high rates of connectivity (migration) among subpopulations (Farias et al. 2010, Santos et al. 2007). In turn, the few studies with cultured tambaquis revealed dramatic reduction in genetic diversity (Calcagnotto and Toledo-Filho 2000, Gomes et al. 2012).
The genetic profile of a fry-producing breeding stock is fundamentally important in fish farming, given the need to avoid potentially deleterious levels of inbreeding, which may eventually have a negative effect on the productivity of farmed stocks. No data are so far available on the genetic variation of the tambaqui breeding stocks used for the production of fry in Brazil, which prevents comparisons with the levels in wild populations. An important question to be examined is whether the genetic variability of the natural population is well represented in the fry breeding stock. The study of Gomes et al. (2012) revealed that Tambaquis farmed in 10 different pisciculture in north Brazil were derived of only two mitochondrial lineages (two females), what is dramatically lower than the haplotype diversity observed in native populations.
The present study was focused in a new area nearby Santarém in central Brazilian Amazonia and aimed to compare the genetic variability of a fry-producing breeding stock, the stocks farmed in this region as well local wild tambaqui populations. The data were compared against the database available in the literature for wild population of Colossoma macropomum of the whole Amazonian base.
MATERIALS AND METHODS
SAMPLES
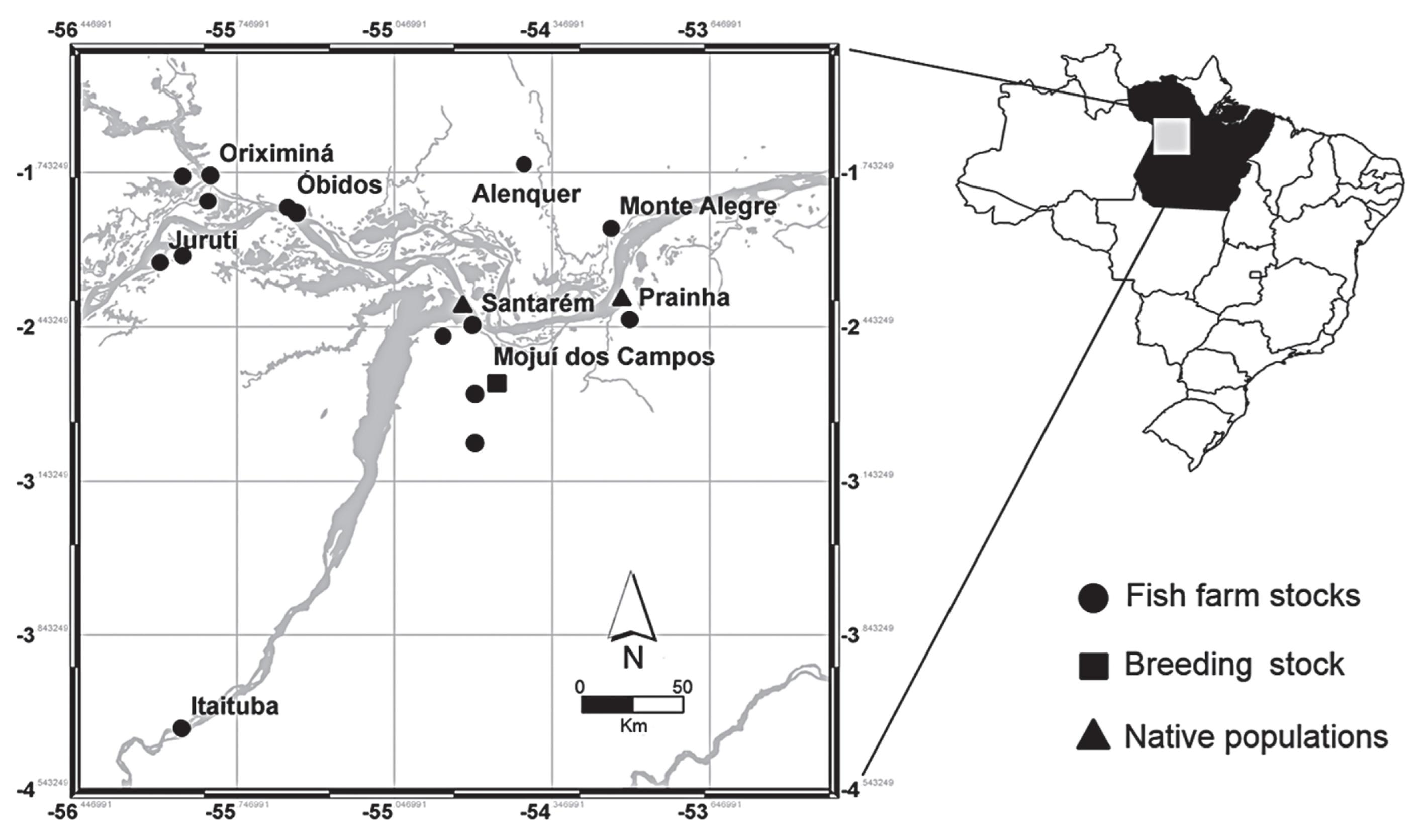
This analysis was based on a sample of 153 individuals collected in the western extreme of the Brazilian state of Pará. Fifty-one specimens were collected in the wild, 27 from the Amazon River near the town of Santarém and 24 from the Curuá-Una River in the town of Prainha. A further 39 specimens were obtained from the fry-producing breeding stock maintained at the Santa Rosa Aquaculture Station in Santarém, and 63 (21 pure C. macropomum, 39 Tambatinga hybrids, and 3 Tambacu hybrids) were collected from 22 fish farms located in municipalities neighboring Santarém (Figure 1). These 63 specimens from pisciculture were previously genotyped by a multiplex of the nuclear Alpha-Tropomyosin gene according to the procedures described by Gomes et al. (2012). All the other specimens from both the wild populations and the fry-producing breeding stock were also analyzed by multiplex and were identified as pure Tambaquis (homozygotes for Tropomyosin).
The specimens obtained from the Santa Rosa Station are part of the total breeding stock of C. macropomum. This facility, which is run by the Pará State Fishery and Aquaculture Ministry (SEPAq), is the main distributor of the fry of tambaqui and other fish species in western Pará. All samples were collected from live specimens, with a small fragment of tissue (approximately 1 cm2) being removed from the tail fin.
Map of the sites at which samples were collected in the wester n extreme of the Brazilian state of Pará
EXTRACTION OF DNA
Total DNA was extracted using Promega's (USA) Wizard(r) Genomic DNA Purification Kit. The protocol involves the lysis of the tissue using the Nuclei Lysis Solution, digestion of proteins with K proteinase, degradation of the RNA with RNase, separation of the proteins with a Protein Precipitation Solution, precipitation and purification of the DNA with isopropanol, washing in ethanol, and hydration of the DNA with Rehydration Solution.
AMPLIFICATION AND SEQUENCING OF THE CONTROL REGION
The Control Region of the mitochondrial DNA was amplified using the primers L1 (5'- CCTAACTCCCAAAGCTAGGTATTC-3') and H2 (5'- TGTTTATCACTGCTGRRTTCCCT-3') described by Santa Brígida et al. (2007). The PCR reactions were prepared in a total volume of 25μl containing: 2.5 μl of buffer (20mM Tris-HCl), 4μl of dNTPs (1.25 mM), 1μl MgCl2 (50 mM), 1.5 μl of DNA, 0.25μl of each primer (L1 and H1, 200 ng/μl), 0.2 μl of Taq polymerase (5U/μl, Invitrogen, Carlsbad, USA), and purified water to complete the final volume of 25 μl. The PCR conditions were: initial denaturation for 3 minutes at 94ºC, followed by 35 denaturation cycles of 30 seconds at 94ºC, hybridization at 57º C for 1 minute, and extension at 72ºC for 2 minutes, with final extension of 10 minutes at 72ºC. The DNA of the products of this process was sequenced using an ABI Prism TM Dye Terminator Cycle Sequencing Reading Reaction kit with electrophoresis in an ABI 3500XL (both from Applied Biosystems, Foster City, USA) sequencer. The new sequences generated in the present study were deposited in GenBank.
DATA ANALYSES
The DNA sequences were edited and aligned in BIOEDIT 5.0.6 (Hall 1999) and a haplotype network was produced using the HaploViewer program (Salzburger et al. 2011). The genetic variability of the population was estimated based on the number of haplotypes (H), haplotype diversity (Nei 1987), and nucleotide diversity (Nei and Tajima 1981). Fu's Fs neutrality test (Fu 1997) was also applied to the data. The distribution of genetic variability within and between populations was inferred from an Analysis of Molecular Variance (AMOVA), complemented with a pairwise analysis of FST values, with significance based on 10,000 random permutations. These indices and parameters of variability and population structure were calculated in Arlequin version 3.5 (Excoffier and Lischer 2010) and DnaSP version 5.10 (Rozas et al. 2003).
RESULTS
A total of 703 base pairs of the mitochondrial Control Region were obtained for each of the 153 specimens, of which 64 sites were polymorphic. In all, 67 haplotypes were recuperated, of which 51 were unique and 16 shared. Seven of these haplotypes were shared between wild and captive specimens. The only haplotype observed in all four populations was H3 (Table I).
Indices of genetic diversity and Fu's Fs para meters are presented in Table II. The wild population have slightly higher levels of both haplotype and nucleotide diversity in comparison with the captive stocks. While haplotype diversity was higher in the fry-producing breeding stock in comparison with the farmed specimens, nucleotide diversity was slightly higher in the latter population. With regard to genetic neutrality (Fs), which estimates the difference between the observed and expected number of haplotypes in wild populations (the captive samples were not included in this analysis), significantly (p < 0.05) negative values were recorded for the two populations: -10.226 for the Amazon and -9.792 for the Curuá-Una.
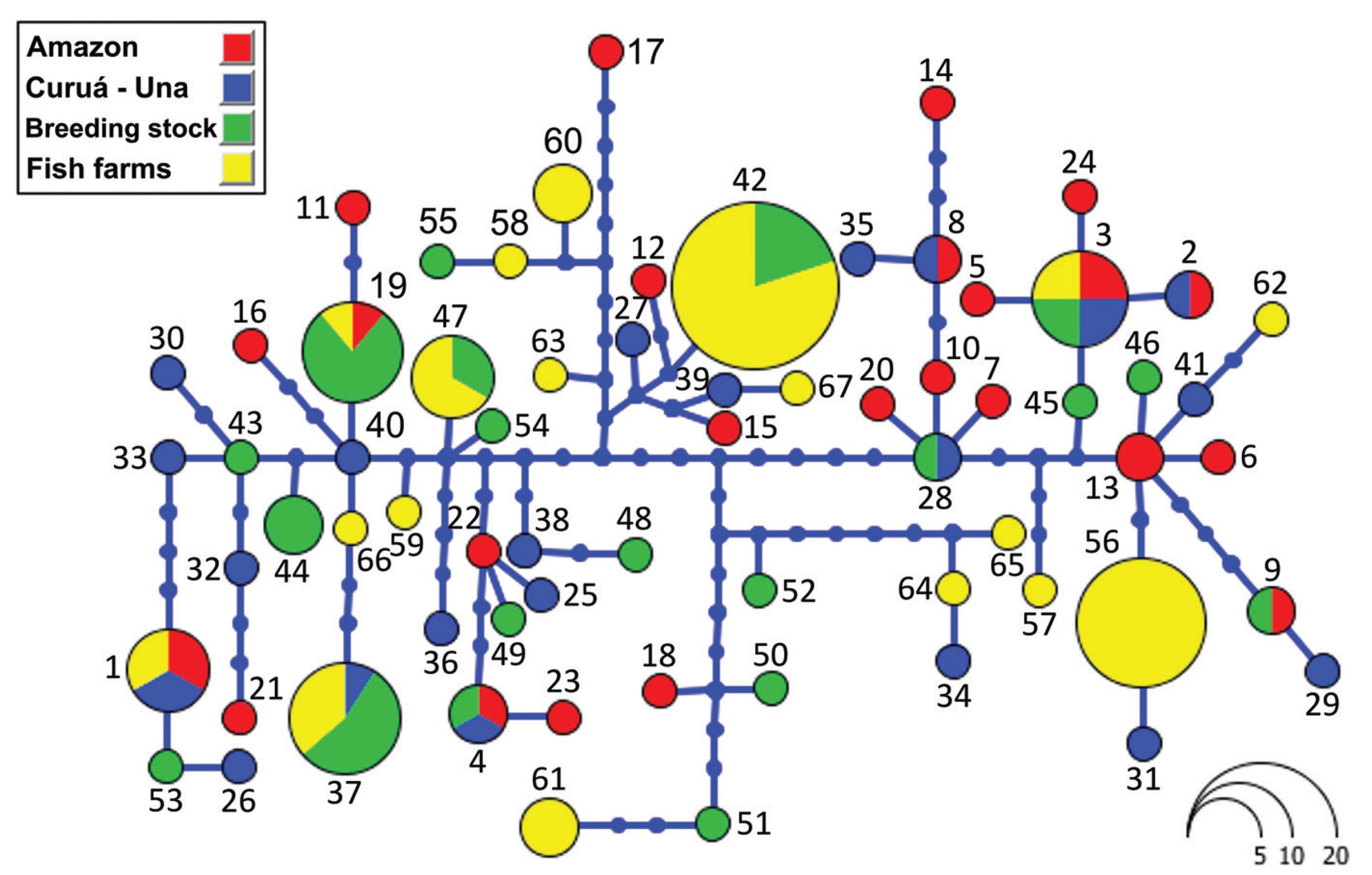
A network haplotype is shown in Figure 2. The haplotypes from the four populations (Amazonas, Curuá-Una, fry-producing breeding stock and farmed stock) appear to be distributed randomly, with no clear geographic pattern or local tendency. As the haplotype diversity of the wild populations is very high (0.9915 and 0.9928), there is a tendency for the specimens from these populations to carry unique haplotypes, whereas in the captive populations, there are more shared haplotypes.
The results of the AMOVA (Table III) indicate the presence of genetic differences in the haplotype composition of the four populations. Most of the genetic variation is found within (93.13%) rather than between (6.87%) populations, but these differences nevertheless result in a highly significant (p < 0.01) Fst value.
Analysis of molecular variance (AMOVA) of the four populations of C. macropomum (Amazon and Curuá-Una rivers, breeding stock, and fish farms) analyzed in the present study.
The pairwise analysis of Fst (Table IV) indicates that the principal differences are between the wild and captive populations, as well as between those of the breeding stock and fish farms. The two wild populations were genetically identical (Fst = 0.00; P = 0.63, not significant), while the breeding stock was somewhat different from the two wild populations (Fst = 0.0954 for the Amazon and 0.03334 for Curuá-Una; P = 0.00391 in both cases), and the fish farm sample (Fst = 0.08938, P = 0.00). There was no statistical difference between the farm population and the wild specimens from the Curuá-Una River (Fst = 0.06982, P = 0.053).
Matrix of pairwise Fst values (bellow the diagonal) as well as Probabilities (above the diagonal) for the comparison between the four study populations of C. macropomum.
DISCUSSION
Aquaculture has been growing worldwide, in particular in response to the increasing demand for food. Despite its positive aspects, this activity poses certain risks, such as potential threats to the genetic integrity of wild populations and inbreeding depression in farmed populations. The Amazonian tambaqui (Colossoma macropomum) is the most popular fish species in Brazil for freshwater aquaculture, and the understanding of the genetic variation should be a priority for the development of management strategies capable of reducing the erosion of the genetic diversity of wild populations and minimizing the risks of inbreeding depression in captive stocks.
Analyzing sequences of the mitochondrial Control Region, the present study found relatively high levels of haplotype diversity in wild populations and surprisingly high values also in samples from a fry-producing stock and fish farms from western Pará state. The haplotype diversity values for the wild populations (0.99) are identical to those recorded by Santos et al. (2007) for wild C. macropomum collected in different locations along the main channel of the Amazonas River (Tabatinga, Coari, Parintins, Oriximina and Santarém) in western and central Amazon basin in Brazil. Our estimation of haplotype diversity for the fry-producing breeding stock (h = 0.93) was only slightly lower than that of the wild populations, and with a large number of haplotypes shared with these populations (see Table II), what indicates that this unit has a quite expressive genetic variability representation of wild populations. This might be expected, up to a point, given that this facility is located on the banks of the Amazon River in Santarém, where the renewal of its stock from the wild is a relatively simple operation.
As expected, the nucleotide diversity (π) of the two wild populations (0.0162 for the Amazon and 0.0168 for the Curuá-Una) was slightly higher than that recorded for the captive populations (0.0155 for the fish farms and 0.0139 for the Santa Rosa stock). These values are very similar to that of 0.012 recorded by Santos et al. (2007). Subsequently, Farias et al. (2010) found similar values in an analysis, which included 127 Tambaquis from Brazil (including the samples of Santos et al), 33 from Peru and 75 from Bolivia. These genetic data on native populations confirm that Colossoma macropomum is a single population in the region studied. Additionally, it is clearly demonstrated by our analysis that the fry-producing stock of Santa Rosa carries a significant representation of the genetic diversity of native populations.
The AMOVA and pairwise Fst comparisons revealed a pattern expected for the wild populations of C. macropomum (Amazon and Curuá-Una), that is, high haplotype diversity, which was also similar to the results of Santos et al. (2007) and Farias et al. (2010) for wild specimens. Neither analytical approach indicated any significant difference between the two wild populations, however. Fu's Fs (Fu 1997) indicates an historic expansion of the population, in other words, the recuperation of the population's genetic variability following an ancient bottleneck, which may not necessarily have been related to the exploitation of the species by modern fisheries.
Comparing the fry-producing breeding stock and the fish farm sample, differences were not accentuated, but were statistically significant in some cases, as it was demonstrated by AMOVA and pairwise Fst. This suggests the loss of haplotypes during the formation of breeding stocks, but possibly is the result of a further loss of diversity during the distribution of the fry to the farms, in particular through the selective sampling of a few breeding females.
The high genetic diversity recorded in the captive specimens analyzed in the present study was unexpected but a good surprise, given the results of a recent study of farmed tambaqui in eastern Pará (Gomes et al. 2012). In marked contrast with the 18 different haplotypes observed in 63 specimens from 22 fish farms in the present study (h = 0.84), Gomes et al recorded only two haplotypes (H1 and H2) for the same mtDNA region in 93 specimens obtained from 10 different fish farms. Interestingly, H1 was recorded in nine of the 10 farms (seven from eastern Pará, one from Amapá, one from Piauí, and one from Rondônia) and H2 was found exclusively in one fish farm located in Santarém, the same area of the present study. This H2 of Gomes et al is 100% identical to haplotype 67 on Figure 2.
Based on the fact that their samples were collected randomly over a two-year period, Gomes et al. (2012) came to the conclusion that haplotype 1 (H1) was historically derived from a single female of C. macropomum, resulting that the principal supplier of fry to most of the farms surveyed, which is located near Castanhal, in eastern Pará, may be affected by a high degree of endogamy resulting from a long history of inbreeding (although it was not possible to confirm this conclusion directly through the analysis of the breeding stock).
These two independent studies focusing in the same species (C. macropomum) indicate the existence of quite distinct scenarios for the farming of the tambaqui and its hybrids in northern Brazil, with some farms raising fry supplied by breeding centers with stocks of very low genetic variability, while others maintain the genetic diversity more typical of wild populations of Colossoma. These two situations appear to represent opposite extremes of the effects of management practices on the genetic structure of the region's farmed stocks.
The data now available for the mitochondrial diversity of wild tambaqui populations appear to support the hypothesis of Santos et al. (2007) and Farias et al. (2010), who concluded that Colossoma macropomum forms a single large panmictic population over its range in the Amazon basin of Brazil, Bolivia, and Peru. This is a potentially important and advantageous characteristic of the species, given that breeding stock can be obtained from the wild in any part of the basin without threatening the genetic viability of captive populations.
Clearly, then, different approaches to the breeding and farming of the species within the same region can have contrasting implications for the genetic diversity of captive stocks. In eastern Pará, management practices lead to the fry-producing stock with reduced genetic variability (Gomes et al. 2012), whereas in the west of the state, the breeding stock with a good representation of wild populations leads to the production of genetically diverse fry (present study). Considering that these two approaches may have both advantages and disadvantages, the data compiled here will be important to guide aquacultural practices that are both economically profitable and ecologically viable to different regions in Brazil. It is especially important to pay attention on genetic impacts on wild populations, in particular through the release of hybrids into the natural environment, whether in the heartland of the distribution of the species - in the Amazon basin - or in areas far from the original range of C. macropomum.
The present study is financed by Universidade Federal do Pará and by Fundação Amazônia Paraense, which provided Jonas Paz Aguiar with a master scholarship. The collection of specimens was authorized by IBAMA through license number 24297. We would like to thank Dr. Lenise Vargas Flôres da Silva and Mr. Zacarias Marques de Oliveira for their support during the collection of specimens at the Santa Rosa station and Davidson Sodre for drawing the map.
REFERENCES
- Araujo-Lima C and Goulding M. 1998. Os frutos do tambaqui. Ecologia, conservação e cultivo na Amazônia. Sociedade Civil Mamirauá/ CNPq/ Rainforest Alliance. Brasília, DF, 186 p.
- Batista VS and Petrere-Jr M. 2003. Characterization of the commercial fish production landed at Manaus, Amazonas state, Brazil. Acta Amazon 33: 53-66.
- Calcagnotto D and Toledo-Filho SDA. 2000. Loss of genetic variability at the transferrin locus in five hatchery stocks of tambaqui (Colossoma macropomum). Genet Mol Biol 23: 127-130.
- Excoffier L and Lischer HEL. 2010. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under linux and windows. Mol Ecol Res 10: 564-567.
- Farias IP, Torrico JP, Garcia-Davila C, Santos MDCF, Hrbek T and Renno J-F. 2010. Are rapids a barrier for floodplain fishes of the amazon basin? A demographic study of the keystone floodplain species Colossoma macropomum (Teleostei: Characiformes). Mol Phylogen Evol 56: 1129-1135.
- Fontes NA, Senhorini JA and Lucas AFB. 1990. Efeito de duas densidades de estocagem no desempenho larval do Paqui, Piaractus mesopotamicus (fêmea x Colossoma macropomum (macho) em viveiros. Bol Tec CEPTA 3: 23-32.
- Fu Y-X. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147: 915-925.
- Gomes F, Schneider H, Barros C, Sampaio D, Hashimoto D, Porto-Foresti F and Sampaio I. 2012. Innovative molecular approach to the identification of Colossoma macropomum and its hybrids. An Acad Bras Cienc 84: 517-526.
- Hall TA. 1999. Bioedit: A user-friendly biological sequence alignment editor and analysis program for windows 95\98\nt. Nucl Ac Symp Ser 41: 95-98.
- Isaac VJ and Ruffino ML. 2003. Informe estatístico do desembarque pesqueiro na cidade de Santarém, PA: 1992-1993. In: Fischer CF (Ed), Recursos pesqueiros do médio amazonas: Biologia e estatística pesqueira IBAMA/GTZ/GOPA, Brasília, p. 225-280.
- Nei M. 1987. Molecular evolutionary genetics Vol. Columbia University Press, New York, 512 p.
- Nei M and Tajima F. 1981. DNA polymorphism detectable by restriction endonucleases Genetics 97: 145-163.
- Ostrensky A, Borghetti JR and Soto D. 2008. Aquicultura no Brasil: O desafio é crescer. Secretaria Especial de Aquicultura e Pesca/Organização das Nações Unidas para Agricultura e Alimentação, Brasília, 276 p.
- Rozas J, Sanchez-Delbarrio JC, Messeguer X and Rozas R. 2003. Dnasp, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496-2497.
- Salzburger W, Ewing GB and Von Haeseler A. 2011. The performance of phylogenetic algorithms in estimating haplotype genealogies with migration. Mol Ecol 20: 1952-1963.
- Santa Brígida EL, Cunha DB, Rego PS, Sampaio I, Schneider H and Vallinoto M. 2007. Population analysis of Scomberomorus cavalla (Cuvier, 1829) (Perciformes, Scombridae) from the northern and northeastern coast of Brazil. Braz J Biol 67: 919-924.
- Santos MCF, Ruffino ML and Farias IP. 2007. High levels of genetic variability and panmixia of the Tambaqui Colossoma macropomum (Cuvier, 1816) in the main channel of the Amazon river. J Fish Biol 71: 33-44.
- Senhorini JA, Figueiredo GM, Fontes NA and Carosfeld J. 1988. Larvicultura e alevinagem do Pacu Piaractus mesopotamicus (Holmberg, 1887), Tambaqui Colossoma macropomum (Curvier, 1818) e seus respectivos híbridos. Bol Tec CEPTA 1: 19-30.
Publication Dates
-
Publication in this collection
21 Oct 2013 -
Date of issue
2013
History
-
Received
07 Jan 2013 -
Accepted
12 Apr 2013