Abstract
Abstract: Candida albicans is the most frequent fungal species that causes infections in humans. Fluconazole is the main antifungal used to treat Candida infections, and its prolonged and indiscriminate use for the last decades are the most established causes which originated resistant strains. Fungal drug resistance is associated to alterations in ERG11 gene and overexpression of multidrug resistance (MDR) transporters belonging to two families: ATP-binding cassette (ABC) and Major Facilitator Superfamily (MFS). To evaluate the role of MFS transporters in azoles resistance of C. albicans clinical strains, this study aimed to analyze four Candida albicans clinical isolates from the University Hospital in Juiz de Fora (Minas Gerais/Brazil), selected in our previous study as they were unaffected by FK506, an ABC pumps inhibitor. In a primary investigation on MFS proteins overexpression, the extrusion of fluorescent substrates (rhodamine 6G and nile red) was analyzed by fluorescence microscopy and flow cytometry. Results suggest participation of MFS transporters in azole resistance of C. albicans isolates and indicate the existence of secondary resistance mechanisms. Therefore, this study contributes to the information about Candida albicans infections in Brazil and reinforces the importance of epidemiological studies focusing on an improved understanding of the disease and further resistance reversion.
Key words
Candida; fluconazole; MFS; multidrug resistance; yeast
INTRODUCTION
Candida species are commensal organisms in healthy individuals. However, in immunocompromised patients, they may cause a disease called candidiasis, which vary from harmless superficial mycosis to highly liLI H, ZHANG C, CHEN Z, SHI W and SUN S. 2014. A promising approach of overcoming the intrinsic resistance of Candida krusei to fluconazole (FLC) - combining tacrolimus with FLC. FEMS Yeast Res 14: 808-811.fe-threatening systemic infections (MorioMORIO F, PAGNIEZ F, LACROIX C, MIEGEVILLE M and LE PAPE P. 2012. Amino acid substitutions in the Candida albicans sterol Delta5,6-desaturase (Erg3p) confer azole resistance: characterization of two novel mutants with impaired virulence. J Antimicrob Chemother 67: 2131-2138. et al. 2012). Fluconazole is the first-line drug for the treatment of Candida infections (Li et al. 2014). Nonetheless, treating candidiasis may be difficult due to the increasing incidence of resistance promoted by its overuse over the past two decades. Moreover, the number of antifungal classes available to treat fungal infections remains extremely limited due to the difficulty of discovering potential drugs that affect the fungus and not the host (DadarDADAR M, TIWARI R, KARTHIK K, CHAKRABORTY S, SHAHALI Y and DHAMA K. 2018. Candida albicans - biology, molecular characterization, pathogenicity, and advances in diagnosis and control - an update. Microb Pathog 117: 128-138. et al. 2018).
The antifungal activity of azoles relies on the inhibition of lanosterol 14α-demethylase (ERG11), a key enzyme in ergosterol biosynthesis. The mechanisms of azole resistance of Candida species are well established, including overexpression or mutations in ERG11 (SanglardSANGLARD D, ISCHER F, KOYMANS L and BILLE J. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14alpha-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother 42: 241-253. et al. 1998); changes in the ergosterol biosynthesis pathway, and energy-dependent drug efflux (Sanglard et al. 1995SANGLARD D, KUCHLER K, ISCHER F, PAGANI JL, MONOD M and BILLE J. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother 39: 2378-2386.). Alterations in ERG11 were considered the primary mechanism of resistance to azoles in C. albicans (KohliKOHLI A, GUPTA V, KRISHNAMURTHY S, HASNAIN SE and PRASAD R. 2001. Specificity of drug transport mediated by CaMDR1: a major facilitator of Candida albicans. J Biosci 26: 333-339. et al. 2001). Nevertheless, the identification of the MDR transporters - ATP binding cassette (ABC) proteins (KaurKAUR R and BACHHAWAT AK. 1999. The yeast multidrug resistance pump, Pdr5p, confers reduced drug resistance in erg mutants of Saccharomyces cerevisiae. Microbiology 145: 809-818. and Bachhawat 1999) and the membrane proteins belonging to the major facilitator superfamily (MFS) (FlingFLING ME, KOPF J, TAMARKIN A, GORMAN JA, SMITH HA and KOLTIN Y. 1991. Analysis of a Candida albicans gene that encodes a novel mechanism for resistance to benomyl and methotrexate. Mol Gen Genet 227: 318-329. et al. 1991) - was soon considered as important as ERG11 modifications on multidrug resistance phenotype (CannonCANNON RD, LAMPING E, HOLMES AR, NIIMI K, BARET PV, KENIYA MV, TANABE K, NIIMI M, GOFFEAU A and MONK BC. 2009. Efflux-mediated antifungal drug resistance. Clin Microbiol Rev 22: 291-321. et al. 2009).
Unlike ABC transporters, dependent on ATP hydrolysis for substrate transport across the membrane, MFS transporters are powered by plasma membrane electrochemical gradient. MDR1, the major MFS transporter gene related to drug resistance (WhiteWHITE TC. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother 41: 1482-1487. 1997), was primarily identified in 1991, in transformed Saccharomyces cerevisiae strains and later, linked to azole resistance in C. albicans (White et al. 1998WHITE TC, MARR KA and BOWDEN RA. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev 11: 382-402.).
Regarding MDR1 gene regulation in C. albicans, it is known that fluconazole does not induce its expression above the normal level, unlike ERG11 (VandeputteVANDEPUTTE PF, FERRARI S and COSTE AT. 2012. Antifungal Resistance and New Strategies to Control Fungal Infections. Int J Microbiol 2012: 26. et al. 2012). However, in many fluconazole-resistant clinical isolates, mutations have probably occurred and abolished the normal regulation of this gene leading to its inherent overexpression in those strains (MorschhauserMORSCHHAUSER J. 2002. The genetic basis of fluconazole resistance development in Candida albicans. Biochim Biophys Acta 1587: 240-248. 2002). This mechanism has already been reported as conferring resistance to different toxic compounds, such as Mdr1p substrates cerulenin and brefeldin A (HillerHILLER D, SANGLARD D and MORSCHHAUSER J. 2006. Overexpression of the MDR1 gene is sufficient to confer increased resistance to toxic compounds in Candida albicans. Antimicrob Agents Chemother 50: 1365-1371. et al. 2006), in a transformed C. albicans strain, and more recently to fluconazole in a clinical isolate (KeniyaKENIYA MV, FLEISCHER E, KLINGER A, CANNON RD and MONK BC. 2015. Inhibitors of the Candida albicans Major Facilitator Superfamily Transporter Mdr1p Responsible for Fluconazole Resistance. PLoS ONE 10: e0126350. et al. 2015), indicating that MFS mediated fluconazole resistance is sufficient to determine if a Candida albicans infection may persist or not.
In Brazil, studies concerning the incidence of Candida infections are decentralized, and the available data regarding fluconazole resistance by membrane transporters is limited (GoulartGOULART LS, SOUZA WWR, VIEIRA CA, LIMA JS, OLINDA RA and ARAUJO C. 2018. Oral colonization by Candida species in HIV-positive patients: association and antifungal susceptibility study. Einstein (São Paulo) 3: eAO4224. et al. 2018). In a previous study, we assessed the frequency of Candida spp. isolated from patients of a Brazilian tertiary hospital, their fluconazole susceptibility and the involvement of ABC transporters on resistance phenotype (Neves-JuniorNEVES-JUNIOR A, CARTAGENES-PINTO AC, ROCHA DA, DE SÁ LF, JUNQUEIRA MDEL and FERREIRA-PEREIRA A. 2015. Prevalence and Fluconazole Susceptibility Profile of Candida spp. Clinical Isolates in a Brazilian Tertiary Hospital in Minas Gerais, Brazil. An Acad Bras Cienc 87: 1349-1359. et al. 2015). From this study, we selected three C. albicans isolates which fluconazole resistance has not been related to ATP-Binding Cassette transporters and evaluated the role of Major Facilitator Superfamily efflux pumps on this process by fluorescence microscopy and flow cytometry.
MATERIALS AND METHODS
YEAST STRAINS AND CULTURE CONDITIONS
Candida albicans clinical isolates were garnered from urine and blood samples of patients in the intensive care unit (ICU) of the University Hospital of Universidade Federal de Juiz de Fora, Minas Gerais, Brazil, between 2012 and 2014. For this study, three resistant strains were selected: 1114, 14A, and 1016; as well as one susceptible strain (134i) and one ABC-mediated fluconazole resistant strain (242A). These C. albicans strains were previously screened as FCZ resistant and were not chemosensitized by FK506, a classic ABC transporter inhibitor (Neves-Junior et al. 2015). It was also used a well-known fluconazole resistant C. albicans clinical isolate (PRI) (Garcia-GomesGARCIA-GOMES AS, CURVELO JA, SOARES RM and FERREIRA-PEREIRA A. 2012. Curcumin acts synergistically with fluconazole to sensitize a clinical isolate of Candida albicans showing a MDR phenotype. Med Mycol 50: 26-32. et al. 2012), and a fluconazole-resistant C. albicans strain (96-25) which overexpresses MDR1 and ERG11, kindly provided by Dr. Theodore C. WhiteWHITE TC, HOLLEMAN S, DY F, MIRELS LF and STEVENS DA. 2002. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob Agents Chemother 46: 1704-1713., University of Washington and Seattle Biomedical Research Institute, Seattle, Washington (White et al. 2002).
This project was approved by Ethics Committee: Protocol CEP-UFJF:079-420-2010 FR:368108; CAEE:0056.420.000-10. All Candida isolates were stored in 20% glycerol at -20°C.
Saccharomyces cerevisiae mutant strains were used as control: one which overexpresses MDR1 efflux pump gene (946), and another with all genes related to the MDR phenotype deleted (522), hypersusceptible to xenobiotic (LampingLAMPING E, MONK BC, NIIMI K, HOLMES AR, TSAO S, TANABE K, NIIMI M, UEHARA Y and CANNON RD. 2007. Characterization of three classes of membrane proteins involved in fungal azole resistance by functional hyperexpression in Saccharomyces cerevisiae. Eukaryot Cell 6(7): 1150-1165. et al. 2007). Both strains were kindly provided by Dr. Brian Monk and Dr. Richard Cannon (University of Otago - New Zealand). A summary containing additional information on the clinical isolates and all control strains used in this study (code, species, site of isolation and strain source) is given in Table I.
All Candida strains were grown in Yeast Extract Peptone Dextrose medium (YPD) (2% glucose, 1% yeast extract, 2% peptone) at 37°C under agitation and were harvested in the exponential growth phase. Saccharomyces strains were grown in the same medium, at 30ºC under agitation.
ANTIFUNGAL AND CHEMICAL AGENTS
Fluconazole (Farmacopa Pharmaceuticals, Rio de Janeiro, Brazil) stock solutions were prepared in distilled water and sterilized by filtration, while itraconazole, voriconazole, posaconazole and ketoconazole (Sigma Aldrich, St. Louis, USA) stock solutions were prepared in dimethyl sulfoxide (DMSO). Stocks were maintained at -20ºC. Rhodamine 6G (Sigma Aldrich, St. Louis, USA) stock solution was prepared in distilled water and stored at room temperature. Nile red (Sigma Aldrich, St. Louis, USA) was prepared in DMSO and maintained at 4ºC.
AZOLES SUSCEPTIBILITY TEST
In vitro azoles susceptibility test was performed and analyzed according to Clinical and Laboratory Standards Institute’s (CLSI) M27-A3 and M27-S4 guidelines (CLSI 2008CLINICAL and LABORATORY STANDARDS INSTITUTE. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts: approved standard M27-A3, 25 p.). Regarding C. albicans, fluconazole’s Minimum Inhibitory Concentration (MIC) end points and classifications for broth microdilution assay are: ≤ 2 μg/ml for susceptible strains; 4μg/ml for susceptible dose-dependent (SDD) strains, and ≥ 8 μg/ml for resistant strains. Itraconazole’s end points are: ≤0.125 µg/ml for susceptible, between 0.25 to 0.5 µg/ml for SDD, and ≥ 1 for resistant. Voriconazole’s end points are: ≤ 0.125 µg/ml for susceptible, between 0.25 to 0.5 µg/ml for SDD, and ≥ 1 µg/ml for resistant. For posaconazole and ketoconazole, the referenced data is not available to indicate a correlation between MIC and outcome of treatment, although MICs for yeast may vary between 0.03 and 16 µg/ml. Cell growth was analyzed in a microplate reader at 600 nm (Fluostar Optima, BMG Labtech, Offenburg, Germany) and compared to the growth control.
RHODAMINE 6G AND NILE RED EFFLUX TESTS IN YEAST CELLS
Efflux methodologies were performed as deDE SÁ LFR et al. 2017. Synthetic Organotellurium Compounds Sensitize Drug-Resistant Candida albicans Clinical Isolates to Fluconazole. Antimicrob Agents Chemother 61: e01231-16.scribed elsewhere (Reis de Sá et al. 2017). Cells at 1 x 10³ cells/ml were incubated overnight in YPD medium, at 37ºC under agitation until the exponential phase of growth was reached. Then, cells were centrifuged (5000 x g /5 min), washed four times with distilled water, and maintained on ice for 2 hours. 6 x 105 cells/ml were incubated with R6G (5 μM) or nile red (7 μM) in phosphate buffered saline (PBS) containing 2% (w/v) glucose, for 1h at 37ºC under agitation. Cells were centrifuged (9000 x g /2 min), washed three times with PBS, observed under a fluorescence optical microscope (Nikon Eclipse E400 microscope - Tokyo, Japan) and analyzed by flow cytometry. For flow cytometry analysis, the experiment was performed in duplicate and analyzed by BD AccuriTM C6 Flow Cytometer. A total of 25.000 cells per sample were measured with a 535nm filter. Flow cytometry data were analyzed using BD Accuri C6 Software (Accuri Cytometers – v. 1.0.264.21). Substrates efflux was evaluated by comparison of mean fluorescence intensity of each strain with control strains and histograms analysis.
STATISTICAL ANALYSIS
Data were presented as mean ± standard error. A probability level of 5% (p < 0.05) in Student’s t-test was considered significant.
RESULTS
STRAINS AND AZOLE SUSCEPTIBILITY PROFILE
In a previous study (Neves-Junior et al. 2015), we demonstrated through a chemosensitization assay, using FK506 (a classical ABC pumps inhibitor), that fluconazole resistance of seven out of 93 Candida strains was not mediated by ABC transporters. Three of the seven C. albicans strains were selected for this study (14A, 1114 and 1016).
Information regarding the susceptibility of the strains to five different azoles (fluconazole, itraconazole, voriconazole, posaconazole, and ketoconazole) are summarized in Table II.. According to CLSI’s breaking points for MIC, 1016, 1114 and 14A strains are resistant to fluconazole. For itraconazole, the three strains were susceptible. 1016 and 1114 strains were susceptible to voriconazole, while 14A displayed a resistance profile. Clinical breakpoints for ketoconazole and posaconazole have not been proposed yet, but they are likely to be close to breakpoints for itraconazole. By using a MIC of ≥ 1 µg/ml to categorize resistant strains, 1016, 1114 and 14A strains could be considered resistant to posaconazole. 1016 and 14A strains could also be considered resistant to ketoconazole, but 1114 should be considered susceptible. PRI strain was included in this study due to the likelihood that its resistance to fluconazole was caused by overexpression of MFS transporters (Neves-Junior et al. 2015). In this study, the PRI strain was categorized as resistant to fluconazole, itraconazole, and voriconazole. A similar susceptibility to azoles was observed for 242A strain, that was resistant to fluconazole, itraconazole, voriconazole, and posaconazole.
Evaluation of the Minimum Inhibitory Concentration (MIC) of azoles against C. albicans and S. cerevisiae strains.
FLUORESCENCE MICROSCOPY ANALYSIS FOR EVALUATION OF ABC AND MFS EFFLUX PUMPS ACTIVITIES
For an initial and qualitative analysis of MFS transporters overexpression in Candida strains, cell fluorescence was observed using a fluorescent optic microscope. In this assay, R6G, an exclusive fluorescent substrate of ABC transporters (HolmesHOLMES AR, LIN YH, NIIMI K, LAMPING E, KENIYA M, NIIMI M, TANABE K, MONK BC and CANNON RD. 2008. ABC transporter Cdr1p contributes more than Cdr2p does to fluconazole efflux in fluconazole-resistant Candida albicans clinical isolates. Antimicrob Agents Chemother 52: 3851-3862. et al. 2008), and nile red, a fluorescent substrate for ABC and MFS transporters (Ivnitski-SteeleIVNITSKI-STEELE I, HOLMES AR, LAMPING E, MONK BC, CANNON RD and SKLAR LA. 2009. Identification of Nile red as a fluorescent substrate of the Candida albicans ATP-binding cassette transporters Cdr1p and Cdr2p and the major facilitator superfamily transporter Mdr1p. Anal Biochem 394: 87-91. et al. 2009), were used as indicators of efflux pumps activity. Cell retention of R6G and extrusion of nile red are the primary characteristics of azole resistance by MFS pumps. Our results are demonstrated in Figure 1 - A columns for R6G and Figure 1 – B columns for nile red. Control mutant strain code 522 was able to retain intracellular fluorescence of both fluorescent substrates. Mutant resistant strain code 946 demonstrated significant nile red extrusion. Similar results were observed on other clinical strains resistant to FCZ (PRI, 1114, 14A, 1016, 96-25). Additionally, all strains were capable of retaining R6G.
Fluorescence microscopy ofC. albicansisolates in the presence of fluorescent substrates, in a 40x magnification. C. albicansstrains PRI, 14A, 1114 and 1016 were treated with rhodamine 6G, an ABC pumps substrate (Acolumns),and nile red, a MFS pumps substrate (Bcolumns). Images show that all strains retained rhodamine 6G. Nonetheless, only strains 522 and 134i retained red nile, while PRI, 1114, 14A, 1016, 946 and 96-25 strains extruded this compound. Saccharomyces cerevisiae strains 522 (mutant strain that lacks efflux pumps) and 946 (mutant strain that overexpresses MFS transporters) were used as controls, as well as C. albicans strains 134i (fluconazole-susceptible clinical isolate) and 96-25 (fluconazole-resistant clinical isolate). Results are representative of at least three independent experiments.
Taken together, the nile red extrusion from the yeast cell and the retention of significant amounts of R6G imply the participation of MFS pumps on azole resistance.
FLOW CYTOMETRY ANALYSIS FOR EFFLUX EVALUATION OF FLUORESCENT SUBSTRATES
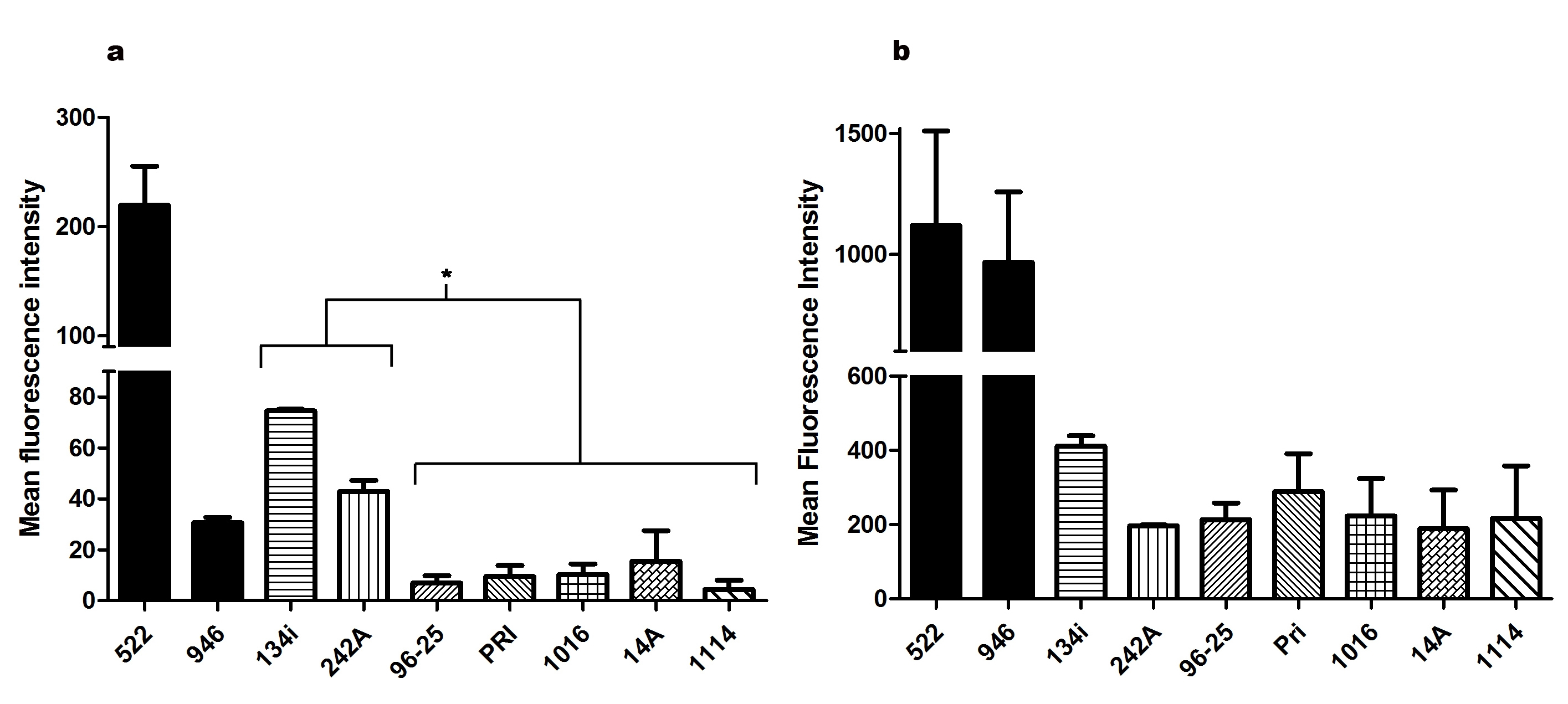
In order to evaluate the efflux pumps activity, cells were loaded with R6G or nile red and their fluorescence intensity was assessed by flow cytometry. Cells were analyzed by the comparison of their geometric mean fluorescence and distribution in a histogram. In nile red efflux analysis (Figure 2a), all strains were able to extrude a significant amount of the compound in comparison to a fluconazole resistant clinical strain, 242A, and a fluconazole susceptible clinical strain, 134i. The C. albicans clinical strain 242A resistance phenotype was associated with ABC transporters overexpression on a previous study using tacrolimus (Neves-Junior et al. 2015). The strains also presented similar efflux rates to 96-25 strain ( ERG11 and MDR1 overexpression) (Figure 2). In the R6G treatment (Figure 2b), 1114, 1016, 14A and PRI presented accumulation of this compound in comparison to 134i strain (susceptible control). Furthermore, histogram analysis showed that only PRI and 1114 strains were able to extrude nile red effectively, although 1114 extruded some of the R6G. Strains 1016 and 14A retained R6G, but little nile red was extruded, indicating a possible secondary resistance mechanism for these two strains, in addition to the MFS transporters participation.
Mean fluorescence intensity ofC. albicansisolates after treatment with nile red or rhodamine 6G. C. albicans strains PRI, 1016, 1114 and 14A were not able to retain nile red, as well as control strain 96-25 (a). The four tested strains retained rhodamine 6G, as well as control strains 134i and 242A (b). Saccharomyces cerevisiaestrains 522 and 946 were used as control. Results are representative of at least four independent experiments. (*) p> 0.05.
DISCUSSION
Even though azole resistance has been a serious problem in therapy for Candida infections for decades (SrivastavaSRIVASTAVA V, SINGLA RK and DUBEY AK. 2018. Emerging virulence, drug resistance and future anti-fungal drugs for Candida pathogens. Curr Top Med Chem 9: 759-778. et al. 2018), this phenomenon still stands as a reality for ICUs and ambulatory institutions around the globe. In Brazilian hospitals, candidemia and invasive candidiasis remain not notified in most of the cases, which leads to difficulties for epidemiologic studies. In Brazil, fluconazole is the first-line therapy in public healthcare institutions for treatment of nearly all Candida infections. Given amphotericin B’s high toxicity, it is only used in critical cases, mostly in systemic infections, and where the first-line treatment has failed. Therefore, the study of azole resistance mechanisms of Candida infections is an important first step to overcome persistent infections and to further analyze the effect of natural of synthetic compounds responsible to revert resistance phenotype.
In this study, we investigated the possible role of MFS efflux pumps in azole resistance of resistant C. albicans clinical isolates. Cross-resistance among azole drugs is expected and observed in hospitals, due to their structural similarity. MIC analysis showed that those strains are resistant to fluconazole and, at least, one other azole. Strains 1016 and 1114 presented an extremely high MIC value for fluconazole (MIC > 1000 µg/ml), but were susceptible to itraconazole and voriconazole. Strain 1114 was also susceptible to ketoconazole. On the other hand, strains 14A and PRI were both resistant to fluconazole (MIC > 64 µg/ml), but with MIC values lower than those observed for strains 1016 and 1114. Interestingly, these strains were resistant to voriconazole and posaconazole. Moreover, strain 14A was resistant to ketoconazole, while strain PRI displayed resistance to itraconazole. These data show that the evaluation of the MIC of antifungal drugs play a pivotal role on the treatment of the patient, since it may guide physicians to prescribe the best therapy according to the susceptibility profile of the etiological agent of the infection.
The approach involving fluorescence evaluation with rhodamine 6G and nile red was previously performed by Garcia-Gomes et al. (2012) in order to analyze efflux pumps activity and suggests possible roles for efflux pumps in yeast resistance (Neves-Junior et al. 2015). Flow cytometry and fluorescence optical microscopy experiments demonstrated the ability of tested C. albicans clinical strains of retaining rhodamine 6G and extruding nile red (Figures 2 and 3). All four strains showed their MFS overexpression potential when a high intracellular accumulation of rhodamine and almost no nile red were observed (Figure 1). Furthermore, the quantitative analysis from flow cytometry indicated the variation of compounds across strains. PRI strain demonstrated the expected pattern for overexpression of MFS transporters. Although 1114 showed a similar profile, this strain also released an amount of R6G for extracellular environment, suggesting the participation of ABC transporters in addition to MFS proteins, unlike the other three strains tested. In the last two strains (14A and 1016), a novel profile of efflux was observed, with much less nile red extrusion. This result may indicate a second resistance mechanism, probably ERG11 related, that could also be responsible for azole resistance, in agreement with MFS pumps overexpression. Previous studies have already demonstrated C. albicans’ ability to persist in an infection by multiple mechanisms such as the overexpression of ABC transporters (ChenCHEN LM, XU YH, ZHOU CL, ZHAO J, LI CY and WANG R. 2010. Overexpression of CDR1 and CDR2 genes plays an important role in fluconazole resistance in Candida albicans with G487T and T916C mutations. J Int Medical Res 38: 536-545. et al. 2010), ERG11 (XiangXIANG MJ, LIU JY, NI PH, WANG S, SHI C, WEI B, NI YX and GE HL. 2013. Erg11 mutations associated with azole resistance in clinical isolates of Candida albicans. FEMS Yeast Res 13: 386-393. et al. 2013), and MFS transporters (CostaCOSTA C, DIAS PJ, SA-CORREIA I and TEIXEIRA MC. 2014. MFS multidrug transporters in pathogenic fungi: do they have real clinical impact? Front Phys 5: 197. et al. 2014).
Flow cytometry histograms of C. albicans isolates after treatment with nile red or rhodamine 6G. In blue: untreated cells; red: cells treated with nile red; black: cells treated with rhodamine 6G. PRI and 1114 extruded a significant amount of nile red and retained rhodamine 6G. In comparison, 1016 and 14A extruded less Nile red and retained rhodamine 6G. These results indicate the possible role of MFS transporters in these strains resistance and suggest a secondary mechanism of resistance for 1016 and 14A. Results are representative of at least four independent experiments.
Data obtained through the fluorescent methodology, however, is limited, since the role of ERG11 cannot be confirmed. This matter justifies our choice of evaluating possible ERG11 participations in resistance by comparisons with genetically modified control strains. By observing the similar efflux profile of the clinical strain which overexpresses ERG11 and MDR1 - 96-25 strain, to the tested strains in flow cytometry, it is unclear whether ERG11 participates in fluconazole resistance of C. albicans isolates, although we may presume the participation of MFS overexpression, since cells were able to extrude nile red.
In summary, this study showed that MFS mediated fluconazole resistance may have an important role in the growth of Brazilian strains at high concentrations of fluconazole. This mechanism of resistance is relevant, since the main clinical impacts of MFS transporters in fungal infections include the development, progression, or persistence of the infection in the host (Costa et al. 2014). Furthermore, in Brazil, there is a lack of information about the molecular mechanisms of azole resistance, as well as the diffuse epidemiological data due to different analysis in different regions of this country (ColomboCOLOMBO AL, NUCCI M, PARK BJ, NOUÉR SA, ARTHINGTON-SKAGGS B, MATTA DA, WARNOCK D and MORGAN J. 2006. Epidemiology of candidemia in Brazil: a nationwide sentinel surveillance of candidemia in eleven medical centers. J Clin Microbiol 44: 2816-2823. et al. 2006). For this reason, this study aims to contribute to the information in Brazil about Candida infections, and also reinforces both the increased reporting of C. albicans azoles resistant strains and the involvement of drug transporters (especially MFS pumps) in this process.
ACKNOWLEGMENTS
The authors are grateful to our lab assistant, Mrs. Geralda Rodrigues Almeida, for her great support and Mr. Eduardo Tessis Castro (University of Toronto, Canada), for corrections and critical review of this manuscript. The authors thank Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for financial support and scholarships.
REFERENCES
- CANNON RD, LAMPING E, HOLMES AR, NIIMI K, BARET PV, KENIYA MV, TANABE K, NIIMI M, GOFFEAU A and MONK BC. 2009. Efflux-mediated antifungal drug resistance. Clin Microbiol Rev 22: 291-321.
- CHEN LM, XU YH, ZHOU CL, ZHAO J, LI CY and WANG R. 2010. Overexpression of CDR1 and CDR2 genes plays an important role in fluconazole resistance in Candida albicans with G487T and T916C mutations. J Int Medical Res 38: 536-545.
- CLINICAL and LABORATORY STANDARDS INSTITUTE. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts: approved standard M27-A3, 25 p.
- COLOMBO AL, NUCCI M, PARK BJ, NOUÉR SA, ARTHINGTON-SKAGGS B, MATTA DA, WARNOCK D and MORGAN J. 2006. Epidemiology of candidemia in Brazil: a nationwide sentinel surveillance of candidemia in eleven medical centers. J Clin Microbiol 44: 2816-2823.
- COSTA C, DIAS PJ, SA-CORREIA I and TEIXEIRA MC. 2014. MFS multidrug transporters in pathogenic fungi: do they have real clinical impact? Front Phys 5: 197.
- DADAR M, TIWARI R, KARTHIK K, CHAKRABORTY S, SHAHALI Y and DHAMA K. 2018. Candida albicans - biology, molecular characterization, pathogenicity, and advances in diagnosis and control - an update. Microb Pathog 117: 128-138.
- DE SÁ LFR et al. 2017. Synthetic Organotellurium Compounds Sensitize Drug-Resistant Candida albicans Clinical Isolates to Fluconazole. Antimicrob Agents Chemother 61: e01231-16.
- FLING ME, KOPF J, TAMARKIN A, GORMAN JA, SMITH HA and KOLTIN Y. 1991. Analysis of a Candida albicans gene that encodes a novel mechanism for resistance to benomyl and methotrexate. Mol Gen Genet 227: 318-329.
- GARCIA-GOMES AS, CURVELO JA, SOARES RM and FERREIRA-PEREIRA A. 2012. Curcumin acts synergistically with fluconazole to sensitize a clinical isolate of Candida albicans showing a MDR phenotype. Med Mycol 50: 26-32.
- GOULART LS, SOUZA WWR, VIEIRA CA, LIMA JS, OLINDA RA and ARAUJO C. 2018. Oral colonization by Candida species in HIV-positive patients: association and antifungal susceptibility study. Einstein (São Paulo) 3: eAO4224.
- HILLER D, SANGLARD D and MORSCHHAUSER J. 2006. Overexpression of the MDR1 gene is sufficient to confer increased resistance to toxic compounds in Candida albicans. Antimicrob Agents Chemother 50: 1365-1371.
- HOLMES AR, LIN YH, NIIMI K, LAMPING E, KENIYA M, NIIMI M, TANABE K, MONK BC and CANNON RD. 2008. ABC transporter Cdr1p contributes more than Cdr2p does to fluconazole efflux in fluconazole-resistant Candida albicans clinical isolates. Antimicrob Agents Chemother 52: 3851-3862.
- IVNITSKI-STEELE I, HOLMES AR, LAMPING E, MONK BC, CANNON RD and SKLAR LA. 2009. Identification of Nile red as a fluorescent substrate of the Candida albicans ATP-binding cassette transporters Cdr1p and Cdr2p and the major facilitator superfamily transporter Mdr1p. Anal Biochem 394: 87-91.
- KAUR R and BACHHAWAT AK. 1999. The yeast multidrug resistance pump, Pdr5p, confers reduced drug resistance in erg mutants of Saccharomyces cerevisiae. Microbiology 145: 809-818.
- KENIYA MV, FLEISCHER E, KLINGER A, CANNON RD and MONK BC. 2015. Inhibitors of the Candida albicans Major Facilitator Superfamily Transporter Mdr1p Responsible for Fluconazole Resistance. PLoS ONE 10: e0126350.
- KOHLI A, GUPTA V, KRISHNAMURTHY S, HASNAIN SE and PRASAD R. 2001. Specificity of drug transport mediated by CaMDR1: a major facilitator of Candida albicans. J Biosci 26: 333-339.
- LAMPING E, MONK BC, NIIMI K, HOLMES AR, TSAO S, TANABE K, NIIMI M, UEHARA Y and CANNON RD. 2007. Characterization of three classes of membrane proteins involved in fungal azole resistance by functional hyperexpression in Saccharomyces cerevisiae. Eukaryot Cell 6(7): 1150-1165.
- LI H, ZHANG C, CHEN Z, SHI W and SUN S. 2014. A promising approach of overcoming the intrinsic resistance of Candida krusei to fluconazole (FLC) - combining tacrolimus with FLC. FEMS Yeast Res 14: 808-811.
- MORIO F, PAGNIEZ F, LACROIX C, MIEGEVILLE M and LE PAPE P. 2012. Amino acid substitutions in the Candida albicans sterol Delta5,6-desaturase (Erg3p) confer azole resistance: characterization of two novel mutants with impaired virulence. J Antimicrob Chemother 67: 2131-2138.
- MORSCHHAUSER J. 2002. The genetic basis of fluconazole resistance development in Candida albicans. Biochim Biophys Acta 1587: 240-248.
- NEVES-JUNIOR A, CARTAGENES-PINTO AC, ROCHA DA, DE SÁ LF, JUNQUEIRA MDEL and FERREIRA-PEREIRA A. 2015. Prevalence and Fluconazole Susceptibility Profile of Candida spp. Clinical Isolates in a Brazilian Tertiary Hospital in Minas Gerais, Brazil. An Acad Bras Cienc 87: 1349-1359.
- SANGLARD D, ISCHER F, KOYMANS L and BILLE J. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14alpha-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother 42: 241-253.
- SANGLARD D, KUCHLER K, ISCHER F, PAGANI JL, MONOD M and BILLE J. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother 39: 2378-2386.
- SRIVASTAVA V, SINGLA RK and DUBEY AK. 2018. Emerging virulence, drug resistance and future anti-fungal drugs for Candida pathogens. Curr Top Med Chem 9: 759-778.
- VANDEPUTTE PF, FERRARI S and COSTE AT. 2012. Antifungal Resistance and New Strategies to Control Fungal Infections. Int J Microbiol 2012: 26.
- WHITE TC. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother 41: 1482-1487.
- WHITE TC, HOLLEMAN S, DY F, MIRELS LF and STEVENS DA. 2002. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob Agents Chemother 46: 1704-1713.
- WHITE TC, MARR KA and BOWDEN RA. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev 11: 382-402.
- XIANG MJ, LIU JY, NI PH, WANG S, SHI C, WEI B, NI YX and GE HL. 2013. Erg11 mutations associated with azole resistance in clinical isolates of Candida albicans. FEMS Yeast Res 13: 386-393.
Publication Dates
-
Publication in this collection
29 July 2019 -
Date of issue
2019
History
-
Received
7 Aug 2018 -
Accepted
13 Nov 2018