Abstract
Abstract: The strains CM-Z19 and CM-Z6, which are capable of highly degrading chlorpyrifos-methyl, were isolated from soil. They were identified as Bacillus megaterium CM-Z19 and Pseudomonas syringae CM-Z6, respectively, based on the 16S rRNA and an analysis of their morphological, physiological and biochemical characteristics. The strain CM-Z19 showed 92.6% degradation of chlorpyrifos-methyl (100 mg/L) within 5 days of incubation, and the strain CM-Z6 was 99.1% under the same conditions. In addition, the degradation characteristics of the two strains were compared and studied, and the results showed that the strain CM-Z19 had higher phosphoesterase activity and ability to degrade the organophosphorus pesticide than did the strain CM-Z6. However, the strain CM-Z19 could not degrade its first hydrolysis metabolite 3,5,6-trichloro-2-pyridinol (TCP) and could not completely degrade chlorpyrifos-methyl. The strain CM-Z6 could effectively degrade TCP and could degrade chlorpyrifos-methyl more quickly than strain CM-Z19.
Key words
Bacillus megaterium; Pseudomonas syringae; chlorpyrifos-methyl; 3; 5; 6-trichloro-2-pyridinol; biodegradation
INTRODUCTION
Chlorpyrifos-methyl (o,o-dimethyl-o-(3,5,6-trichloro-2-pyridyl) phosphorothioate, also called CM), is a broad-spectrum organophosphorus insecticide that has been widely used in the prevention of agriculture pests since 1975 (SzpyrkaSZPYRKA E, MATYASZEK A and SŁOWIK-BOROWIEC M. 2017. Dissipation of chlorantraniliprole, chlorpyrifos-methyl and indoxacarb-insecticides used to control codling moth (Cydia pomonella, L.) and leafrollers (Tortricidae) in apples for production of baby food. Environ Sci Pollut R 24(13): 12128-12135. et al. 2017, CHO KM, MATH RK, ISLAM SM, LIM WJ, HONG SY, KIM JM, YUN MG,CHO JJ and YUN HD. 2009. Biodegradation of chlorpyrifos by lactic acid bacteria during kimchi fermentation. J Agric Food Chem 57(5): 1882-1889. ChoiCHOI WS and LEE SE. 2016. Toxicity of chlorpyrifos-methyl to Sitophilus zeamais collected in korea and biochemical differences. Entomol Res 46(1): 15-22. et al. 2016). The excessive and frequent application of CM may result in high levels of CM residues on agricultural products, soil and water, which might have adverse effects on environmental health and public health (MatthewsMATTHEWS WA. 2010. The fate of chlorpyrifos-methyl in stored wheat: a comparison of a laboratory-scale experiment with a pilot-scale treatment. Pest Manag Sci 30(1): 21-29. et al. 2010, AbbassyABBASSY MA, SALIM YMM, SHAWIR MS and NASSAR AMK. 2017. Disappearance and hazard quotient of chlorpyrifos-methyl, fipronil, and imidacloprid insecticides from dates. J Consum Prot Food S 12(3): 223-230. et al. 2017). These problems highlight the need to remove CM residues from contaminated soils, water systems, and crops. Microbial degradation has been considered the most important bioremediation method, because it is the main factor impacting CM degradation (KimKIM JR and AHN YJ. 2009. Identification and characterization of chlorpyrifos-methyl and 3,5,6-trichloro-2-pyridinol degrading Burkholderia sp. strain KR100. Biodegradation 20(4): 487-497. and Ahn 2009). The most common degradation pathway of CM involves the formation of 3,5,6-trichloro-2-pyridinol (TCP) by microorganisms, and TCP is a more hazardous pollutant than the parent compound (Kim et al. 2009, BlanchetBLANCHET P and ST-GEORGE A. 2010. Kinetics of chemical degradation of organophosphorus pesticides; hydrolysis of chlorpyrifos and chlorpyrifos-methyl in the presence of copper(ii). Pest Manag Sci 13(1): 85-91. and St-George. 2010). TCP is more mobile in soil and more leachable into groundwater and surface water (AbrahamABRAHAM J and SILAMBARASAN S. 2013. Biodegradation of chlorpyrifos and its hydrolyzing metabolite 3,5,6-trichloro-2-pyridinol by Sphingobacterium sp. JAS3. Process Biochem 48(10): 1559-1564. and Silambarasan 2013, TitusTITUS KR. 1988. Degradation of chlorpyrifos and its hydrolysis product, 3,5,6-trichloro-2-pyridinol, in soil. J Environ Sci Heal B 23(6): 527-539. et al. 1988), and it causes widespread contamination in the soils and in the aquatic environment (Abraham and Silambarasan 2016ABRAHAM J and SILAMBARASAN S. 2016. Biodegradation of chlorpyrifos and its hydrolysis product 3,5,6-trichloro-2-pyridinol using a novel bacterium Ochrobactrum sp. JAS2: a proposal of its metabolic pathway. Pestic Biochem Physiol 126(1): 13-21.). The half-life of TCP ranges from 65 to 360 days in soil and is considered to be a persistent pollutant by US EPA (MayaMAYA K, UPADHYAY SN, SINGH RS and DUBEY SK. 2012. Degradation kinetics of chlorpyrifos and 3,5,6-trichloro-2-pyridinol (TCP) by fungal communities. Bioresour Technol 126: 216-223. et al. 2012). It was suggested that the accumulation of TCP, which has anti-microbial properties, prevents the proliferation of CM-degrading microorganisms (FinocchiaroFINOCCHIARO R, MELI S and GENNARI M. 2004. Behavior of chlorpyrifos-methyl in soil and sediment. J Environ Sci Heal B 39(3): 381-392. et al. 2004, AnwarANWAR S, LIAQUAT F, KHAN QM, KHALID ZM and IQBAL S. 2009. Biodegradation of chlorpyrifos and its hydrolysis product 3,5,6-trichloro-2-pyridinol by Bacillus pumilus strain C2A1. J Hazard Mater 168(1): 400-405. et al. 2009). Therefore, the biodegradation of CM concerns not only the concentration of CM but also its intermediate metabolites, especially TCP.
At present, isolation and application of CM-degrading bacteria is considered to be an effective way to treat CM contamination (Kim and Ahn 2009). A small number of CM-degrading bacteria have been isolated from the environment, but studies of these bacteria are usually only concerned with the concentration of CM in the biodegradation process, and there is minimal understanding of the degradation intermediates and degradation mechanisms. In this study, two CM-degrading bacteria (Bacillus megaterium CM-Z19 and Pseudomonas syringae CM-Z6) were isolated from soils, and their degradation potential and degradation characteristics were studied. The main purpose of this study was to better understand the mechanism of microbial degradation of CM, thereby laying a foundation for the rational use and management of CM and related pesticides. In addition, this study might also provide a foundation for the application of Bacillus megaterium CM-Z19 and Pseudomonas syringae CM-Z6.
MATERIALS AND METHODS
CHEMICALS AND MEDIA
Chemicals: CM (99.5%) was purchased from Sigma-Aldrich. 3,5,6-trichloro-2-pyridinol (94.0%) was purchased from Dr. Ehrenstorfer GmbH. All other chemicals used were of analytical grade and were commercially available.
Culture media: (1). Inorganic salt medium: MgSO4·7H2O 0.2 g, (NH4)2SO4 0.1 g, K2HPO4 0.1 g, CaSO4 0.05 g, FeSO4·7H2O 0.01 g, water 1 L, pH 7.0. (2). Isolation medium: inorganic salt medium containing 100 mg/L CM was used to isolate CM-degrading bacteria. (3). Enrichment medium: peptone 10 g, yeast extract powder 5 g, beef extract powder 3 g, sodium chloride 5 g, water 1 L, pH 7.0.
ISOLATION AND IDENTIFICATION OF CM-DEGRADING BACTERIA
The soil sample was collected from a farmland in ZhenjiJI X, WANG Q, ZHANG W and YIN F. 2016. Research advances in organophosphorus pesticide degradation: a review. Fresen Environ Bull 25(5): 1556-1561.ang City, China, in which CM has been used for more than 3 years. The 3.0 g soil sample was placed in 100 ml isolation medium for shaking culture at 30°C (120 rpm). The concentration of the residual CM was determined every 24 h (PengPENG G, LU Y, HE Q, MMEREKI D, ZHOU G, CHEN J and TANG X. 2016. Determination of 3,5,6-trichloro-2-pyridinol, phoxim and chlorpyrifos-methyl in water samples using a new pretreatment method coupled with high-performance liquid chromatography. J Sep Sci 38(24): 4204-4210. et al. 2016). The 5 ml culture solution with 3-d degradation rate >70% was transferred to an enrichment medium containing same concentration of CM, and then continuously transferred more than 5 times. After the degradation effect was verified again, the above enrichment medium was coated on an inorganic salt culture plate containing CM for an inverted culture at 30°C. The eugonic bacterial colonies were selected and repeatedly streaked on the culture plate to obtain the pure culture.
The new isolates were identified by using the 16S rRNA sequence analysis. Total DNA was extracted following the standard procedure (SambrookSAMBROOK J and RUSSELL DW. 1998. Molecular cloning: a laboratory manual. 3rd ed., Cold Spring Harbor Laboratory Press. and Russell 1998, XiaXIA M, HUAN C, JIANG T, ZHENG Z and OUYANG J. 2017. Molecular cloning and enzymological characteristics of a novel N-acetyl-β-D-glucosaminidase from Bacillus coagulans. J Forest Eng 2(2): 70-75. et al. 2017). Nucleotide primers 27F (5’-AGAGTTTGATCCTGGTCAG-3’) and 1492R (5’-TACGGCTACCTTGTTACGACT-3’) were used to amplify the 16S rDNA (ShiSHI XZ, GUO RJ, TAKAGI K, MIAO ZQ and LI SD. 2011. Chlorothalonil degradation by Ochrobactrum lupini strain TP-D1 and identification of its metabolites. World J Microb Biotech 27(8): 1755-1764. et al. 2011). The PCR reaction was performed in a PCR thermocycle instrument with the following cyclic profile: initial denaturation at 94°C for 5 min, 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, extension at 72°C for 2 min and final extension at 72°C for 10 min (Cho et al. 2009). The determined 16S rRNA gene (partial sequence) was aligned with those available in the GenBank database. A phylogenetic tree was made by using ClustalX1.83 and MEGA 5.0 (WangWANG S, ZHANG C and YAN Y. 2012. Biodegradation of methyl parathion and p-nitrophenol by a newly isolated Agrobacterium sp. strain Yw12. Biodegradation 23(1): 107-116. et al. 2012, ZengZENG X. 2015. Identification of the pathogen causing Eucalyptus anthrax and laboratory screening of fungicides. China Forest Sci Technol (J Forest Eng) 29(4): 121-125. 2015).
DEGRADATION OF CM BY STRAIN CM-Z6 AND STRAIN CM-Z19
Inoculums of CM-degrading bacteria were prepared in advance. The bacteria were grown in a nutrient broth medium at 30°C, 100 rpm. Cells were collected after overnight culture by centrifugation and washed with the sterilized saline. Approximately 2×105 cells/ml were resuspended in saline. This bacterial suspension will be used as inoculum for subsequent experiments (ZhuZHU J, ZHAO Y, XIE K, LI X and CHEN Y. 2018a. Study on biodegradation of chlorpyrifos and utilization of a chlorpyrifos degrading bacterium, Bacillus amyloliquefaciens CZ-5. Fresen Environ Bull 27(2): 1196-1201. et al. 2018a).
A 4% inoculum of strain CM-Z6 was inoculated in inorganic salt culture medium containing 100 mg/L CM, and then was placed in the incubator (120 rpm, 30°C). The concentration of CM and TCP were determined every 24 h (Peng et al. 2016). The strain CM-Z19 was tested synchronously as described above.
DEGRADATION OF TCP BY STRAIN CM-Z6 AND STRAIN CM-Z19
The inorganic salt culture medium containing 100 mg/L TCP was inoculated with a 4% inoculum of strain CM-Z6, and then was placed in the incubator (120 rpm, 30°C). The concentration of TCP was determined regularly. The strain CM-Z19 was tested synchronously in the same manner.
DEGRADATION OF 7 ORGANOPHOSPHORUS PESTICIDES BY STRAIN CM-Z6 AND STRAIN CM-Z19
The sterile soil was divided into 7 groups. Group 1: Sterile triazophos was added to the soil at a concentration of 100 mg/kg, and then soil was inoculated with strain CM-Z6. The inoculation amount was 10 ml of inoculum per 100 g of soil; finally, the culture was carried out in the incubator. The water-holding capacity of the soil was adjusted to 40% during the incubation period. The residual concentration of the pesticide was measured at intervals of 48 h (ProdhanPRODHAN MDH, PAPADAKIS EN and PAPADOPOULOU-MOURKIDOU E. 2015. Determination of multiple pesticide residues in eggplant with liquid chromatography-mass spectrometry. Food Anal Method 8(1): 229-235. et al. 2015). The effect of the strain CM-Z6 on the degradation of the organophosphorus pesticide in soil was evaluated. The experimental pesticides in Groups 2 to 7 were changed to phorate, monocrotophos, methyl parathion, fenitrothion, parathion and isocarbophos, respectively. The degradation of 7 organophosphorus pesticides by strain CM-Z19 was tested in the same manner.
PHOSPHOESTERASE ACTIVITY OF THE STRAINS CM-Z6 AND CM-Z19
To 100 ml CM-Z6 (or CM-Z19) culture medium, a protease inhibitor mixture was added, and the medium was centrifuged at 10,000 r/min for 5 min, while the precipitate (α) and supernatant (I) were collected separately. The precipitate (α) was washed twice with 50 mmol/L Tris-HCl buffer, and the wash solution was added to the supernatant (I), and freeze-dried to near dryness, make up to a constant volume of 20 ml for testing extracellular enzyme activity. The precipitate (α) was set to 20 ml with Tris-HCl buffer, to which the appropriate amount of protease inhibitor was added, and the sample was sonicated in an ice bath (200 W, broken for 5 s, intervals of 4 s, total 4 min) and then centrifuged at 10,000 r/min for 10 min. The supernatant (II) contained the crude enzyme solution (set to 20 ml to determine the enzyme activity). The crude enzyme solution was centrifuged at 18,000 r/min for 30 min, and then the supernatant (β) was resuspended in 20 ml Tris-HCl buffer to assay membrane-bound enzymes. Supernatant (III) was reset to 20 ml with Tris-HCl buffer for intracellular enzyme assays (DasDAS S and SINGH DK. 2006. Purification and characterization of phosphotriesterases from Pseudomonas aeruginosa F10B and Clavibacter michiganense subsp. insidiosum SBL11. Can J Microbiol 52(2): 157-168. and Singh 2006, BhadbhadeBHADBHADE BJ, SARNAIK SS and KANEKAR PP. 2002. Biomineralization of an organophosphorus pesticide, monocrotophos, by soil bacteria. J Appl Microbiol 93(2): 224-234. et al. 2002).
The phosphoesterase enzymatic reaction system (3 ml) was as follows: 2.8 ml glycine-sodium hydroxide buffer (0.05 mol/L, pH 9.0), 0.19 ml of enzyme solution, 10 μl of ρ-nitrophenyl phosphate disodium salt (40 mmol/L), 37°C water bath for 1 h. Reaction solution (0.5 ml) was added into the test tube, with 3.5 ml glycine-sodium hydroxide buffer (pH 10.0), and OD405 was used to determine the amount of p-nitrophenol in solution (colorimetric determination). A unit of enzyme activity (U) was defined as the amount of enzyme required to convert ρ-nitrophenyl phosphate disodium salt into 1 μmol p-nitrophenol in 1 min.
RESULTS AND DISCUSSION
IDENTIFICATION AND CHARACTERIZATION OF CM-DEGRADING BACTERIA
Two bacteria, strains CM-Z6 and CM-Z19, that are capable of degrading CM were isolated from the soil. The 3-d degradation rates of CM (100 mg/L) for CM-Z6 and CM-Z19 were 88.8% and 71.3%, respectively. The strain CM-Z6 was gram-negative, aerobiotic, mobile, non-sporous, rod-shaped, and 0.6-0.8×1.5-2.5 µm in size, and it formed circular and wet colonies on the nutrient agar plates. It was positive for catalase, oxidase and gelatin liquefaction, however negative for the denitrification test, VP test and indole test (SangkharakSANGKHARAK K and PRASERTSAN P. 2012. Screening and identification of polyhydroxyalkanoates producing bacteria and biochemical characterization of their possible application. J Gen Appl Microbiol 58(3): 173-182. and Prasertsan 2012, SongSONG X, ZHANG Z, LIU Y, ZHAN N and XIANG D. 2016. Isolation and identification of microorganisms causing mildew of fiberboard in Nanning,Guangxi. J Forest Eng 1(1): 78-82. et al. 2016, ZhuZHU J, ZHAO Y, LI X, CHEN W and HUANG R. 2018b. Study on the characteristics and utilization of a triclopyr butoxyethyl ester degrading bacterium, Lactobacillus buchneri TBE-6. Fresen Environ Bull 27(5): 3156-3161. et al. 2018b). The strain CM-Z19 was a straight or circulate bacillus, gram-positive, aerobiotic, mobile, spore-forming, and 0.6-0.7×2.0-2.5 µm in size, and it formed white, opaque, rough colonies on the nutrient agar plate. It was positive for catalase, oxidase, starch hydrolysis, gelatin liquefaction and the VP test, but negative for the indole test.
The 16S rDNA sequences that were obtained by PCR amplification of the strains CM-Z6 and CM-Z19 were 1411 bp and 1406 bp, respectively (GenBank Accession no. KY038875 and KY038876). Blastn of the 16S rDNA sequence indicated that the strains CM-Z6 and CM-Z19 showed nearly 99% similarity with Pseudomonas syringae and Bacillus megaterium, respectively. The phylogenetic trees of CM-Z6 and CM-Z19 are shown in Figs. 1 and 2.
DEGRADATION OF CM BY STRAIN CM-Z6 AND BY STRAIN CM-Z19
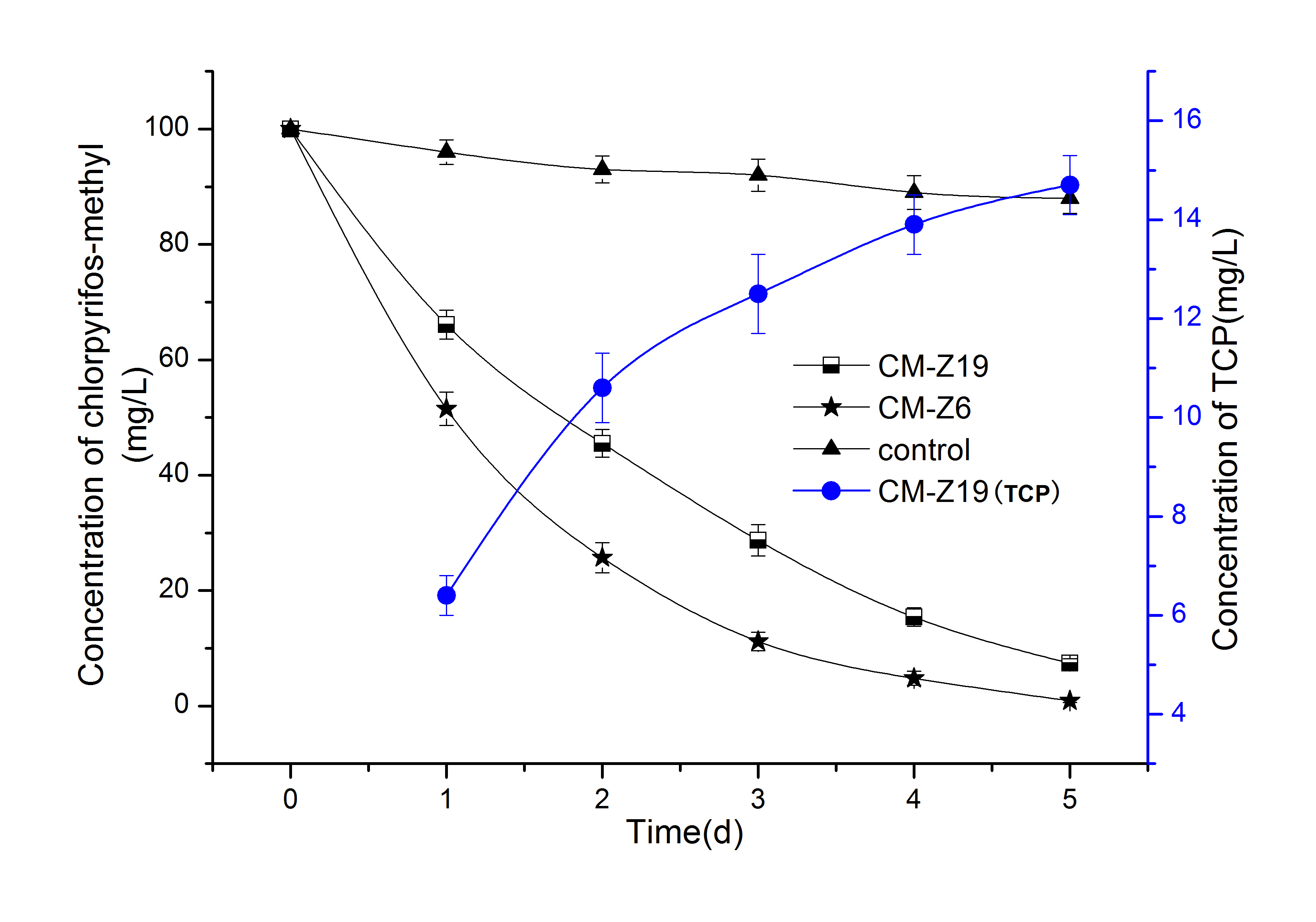
Fig. 3 shows the degradation curves of CM by strain CM-Z6 and by strain CM-Z19, as well as the concentration of TCP generated during degradation. The results showed that both strains CM-Z6 and CM-Z19 could effectively degrade CM, with 3-d degradation rates of 88.8% and 71.3%, respectively, which is in line with the data reported by MoscosoMOSCOSO F, TEIJIZ I, DEIVE FJ and SANROMÁN MA. 2013. Approaching chlorpyrifos bioelimination at bench scale bioreactor. Bioproc Biosyst Eng 36(9): 1303-1309. et al. 2013, who studied various bacteria involved in the degradation of chlorpyrifos and chlorpyrifos-methyl and proposed a higher degradation ability for Pseudomonas than for Bacillus (Moscoso et al. 2013, MayaMAYA K, SINGH RS, UPADHYAY SN and DUBEY SK. 2011. Kinetic analysis reveals bacterial efficacy for biodegradation of chlorpyrifos and its hydrolyzing metabolite TCP. Process Biochem 46(11): 2130-2136. et al. 2011). TCP is one of the most important intermediates in the process of CM degradation (Kim and Ahn 2009). Typically, a phosphoesterase hydrolyzes CM to TCP. Both strains CM-Z6 and CM-Z19 have shown phosphoesterase activity (Fig.7). Therefore, it was inferred that TCP should be produced in the degradation of CM by the two strains. However, the results in Fig. 3 show that TCP was detected only during the degradation of CM by strain CM-Z19, and no TCP was detected during the degradation of CM by strain CM-Z6 (the TCP of the control group was also 0). In the process of degradation of CM by strain CM-Z19, the concentration of TCP in the culture medium gradually increased with the gradual degradation of CM (Fig. 3). Therefore, it was speculated that TCP produced during the degradation of CM by strain CM-Z19 could not be degraded or very slowly.
The results in Fig. 4 show that the strain CM-Z19 could not degrade TCP (no significant difference with the control group, P> 0.05), and the strain CM-Z6 could effectively degrade TCP. The degradation rate of TCP by strain CM-Z6 reached approximately 85% in 48 h, and it was significantly higher than that of Ochrobactrum sp. JAS2 and Bacillus pumilus C2A1 (Abraham and Silambarasan 2016, Anwar et al. 2009).
In the degradation of CM by strain CM-Z6, the reason why no TCP was detected may be due to the rapid degradation of the generated TCP. The statistical analysis of the experimental results in Fig. 3 shows that the degradation curve of CM by strain CM-Z6 conforms to a first-order kinetics model, and the fitted equation was C=100e-1.2846t, R2=0.9921, half-life=0.95 d. The amount of degraded CM could be considered to be the maximum amount of TCP that could be generated (in molar concentration). It had been confirmed that strain CM-Z6 can rapidly degrade TCP (Fig. 4) and can grow by using TCP as the sole carbon source. The fitted equation for the degradation of TCP was C=100e-1.5493t,R2=0.9795,half-life=0.92 d. Comparing the above two groups of degradation parameters, it could be that the rate constant of TCP degradation was higher than that of CM and it had a shorter half-life. Therefore, in the process of degradation of CM by strain CM-Z6, TCP as the intermediate would not accumulate, and no TCP would be detected. In addition, the degradation of TCP could help to reduce feedback suppression and promote the degradation of CM (MeikleMEIKLE RW and YOUNGSON CR. 1978. The hydrolysis rate of chlorpyrifos, o-o-diethyl o-(3,5,6-trichloro-2-pyridyl) phosphorothioate, and its dimethyl analog, chlorpyrifos-methyl, in dilute aqueous solution. Arch Environ Con Tox 7(1): 13-22. and Youngson 1978, LiLI C, LIU H, HAO Z, KE H, TAO G and NI H. 2012. Characterization of a newly isolated highly effective 3,5,6-trichloro-2-pyridinol degrading strain Cupriavidus pauculus P2. Curr Microbiol 65(3): 231-236. et al. 2012, SilambarasanSILAMBARASAN S and ABRAHAM J. 2014. Efficacy of Ganoderma sp. JAS4 in bioremediation of chlorpyrifos and its hydrolyzing metabolite TCP from agricultural soil. J Basic Microb 54(1): 44-55. and Abraham 2014).
DEGRADATION OF 7 ORGANOPHOSPHORUS PESTICIDES BY THE STRAINS CM-Z6 AND CM-Z19
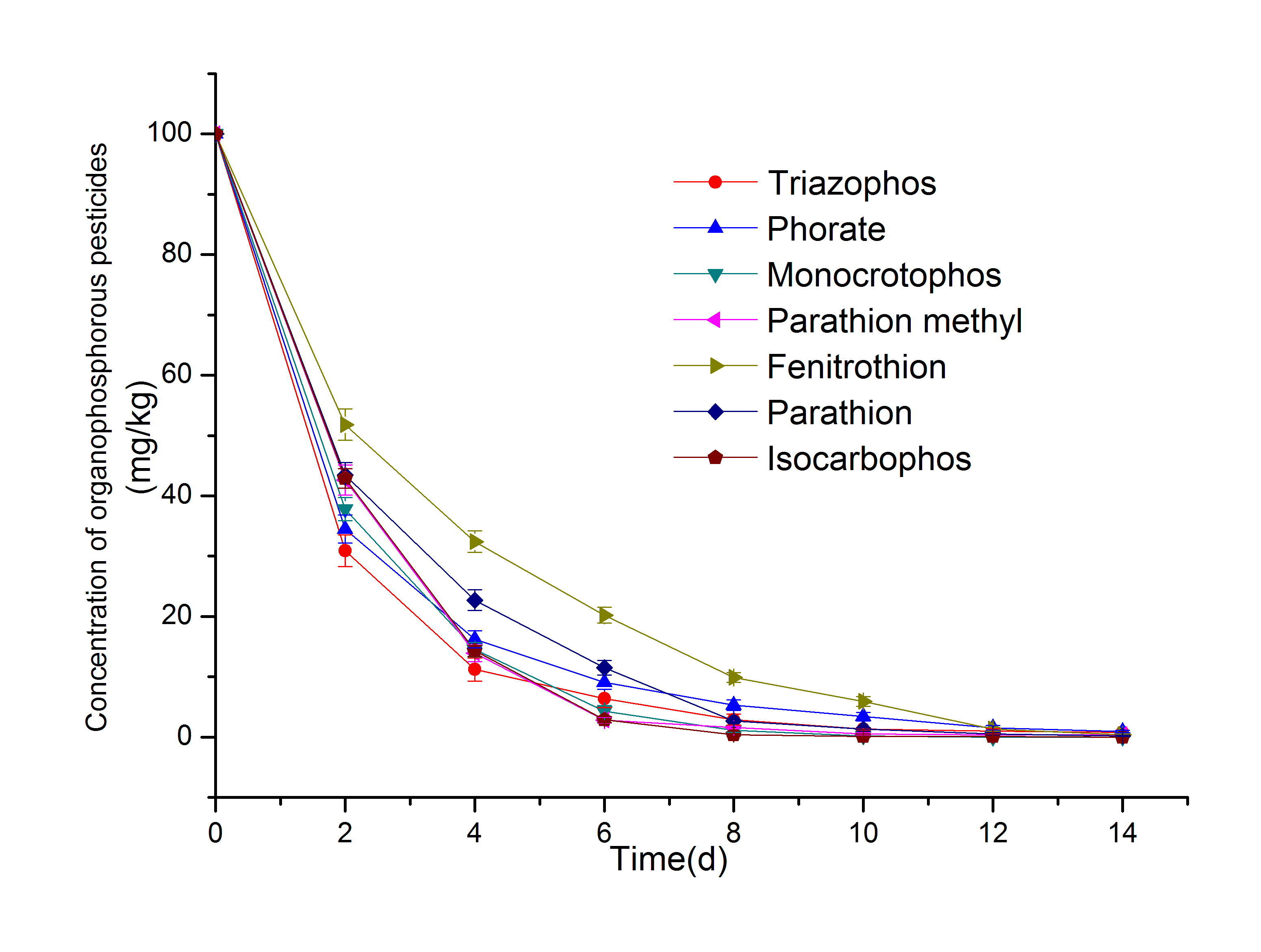
The results in Fig. 5 show that strain CM-Z19 had an excellent degradation effect on 7 organophosphorus pesticides, and the 8-d degradation rate was more than 90%, and the 14-d degradation rate was higher than 99%. These results indicated that the degradative effect of strain CM-Z19 on organophosphorus pesticides was broad, and the reason may be that the degrading enzymes of strain CM-Z19 can act on the common structure of organophosphorus pesticides. Organophosphorus pesticides generally have a common structure with that of phosphates (or phosphorothioates or phosphoramidates), and microorganisms that degrade such common structures can usually degrade a variety of organophosphorus pesticides. The rate of degradation may be related to the chemical stability of the pesticide itself, the spatial structure, and the feedback effects of the pesticide and the intermediate metabolites on the enzymatic reaction (LeeLEE SE. 2011. Purification and characterisation of a carboxylesterase from a chlorpyrifos-methyl-resistant strain of Oryzaephilus surinamensis (Coleoptera: Silvanidae). Aust J Entomol 50(2): 187-194. 2011, Ji et al. 2016, Zhu et al. 2019ZHU J, FU L, JIN C, MENG Z and YANG N. 2019. Study on the isolation of two atrazine-degrading bacteria and the development of a microbial agent. Microorganisms 7(3): 80.).
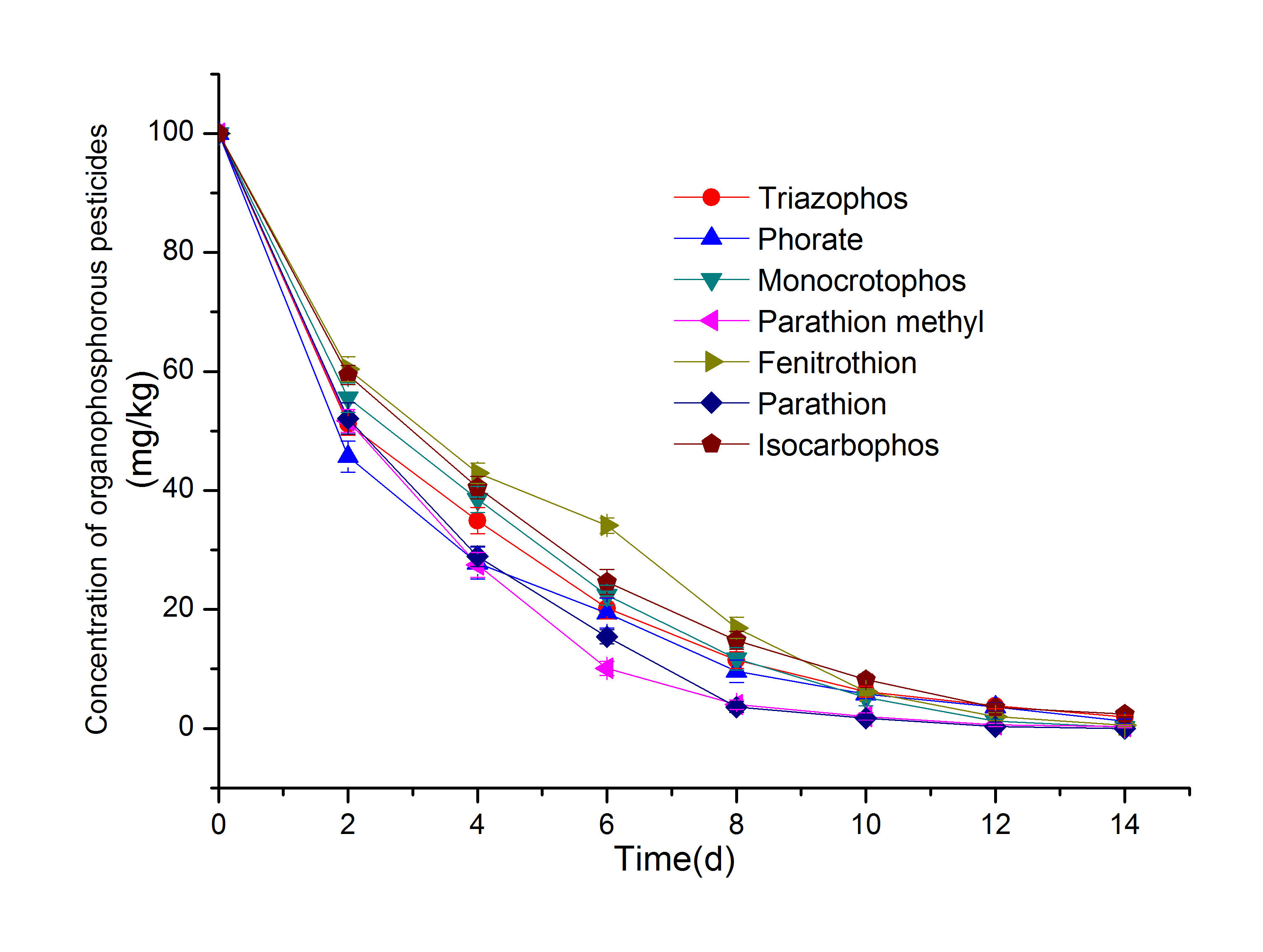
In addition, the degradation of 7 organophosphorus pesticides by Pseudomonas syringae CM-Z6 is shown in Fig. 6, the 8-d degradation rate was 83% or higher. However, the comparative analysis of the results in Figs. 5 and 6 shows that the degradation rate of 7 organophosphorus pesticides by strain CM-Z19 was higher than that of strain CM-Z6. This was inconsistent with the results for the degradation of CM (the results of previous experiments have shown that the degradation rate of CM by strain CM-Z6 is significantly higher than that of strain CM-Z19). This result may be closely related to the type and activity of organophosphorus-degrading enzymes that are produced by microorganisms.
Microorganisms that are capable of degrading organophosphorus pesticides may produce a variety of organophosphorus pesticide-degrading enzymes; one of the most important categories is phosphoesterase. In general, phosphoesterases can hydrolyze the phosphate ester bond of most organophosphorus pesticides. If microorganisms could produce such enzymes that could act on the common structure of organophosphorus pesticides, their degradative effect on organophosphorus pesticides would be broad.
In this study, the ability of strain CM-Z19 to degrade organophosphorus pesticides was better than that of strain CM-Z6 as a whole, probably due to the higher activity of the degrading enzyme that is produced by strain CM-Z19. The ability of strain CM-Z19 to degrade CM was not as good as that of strain CM-Z6, which may be because strain CM-Z6 produces specific enzymes that either directly lead to the degradation of CM or decompose the intermediates of CM (e.g., TCP), which would be conducive to the rapid progress of the degradation reaction. The capacity for TCP utilization by strain CM-Z6 could be the reason that this isolate was more efficient than others in its ability to degrade CM and survive better in a culture medium, whereas, the inability to degrade an unknown metabolite or TCP could be the reason for the lesser degree of CM degradation shown by strain CM-Z19 (LakshmiLAKSHMI CV, KUMAR M and KHANNA S. 2009. Biodegradation of chlorpyrifos in soil by enriched cultures. Curr Microbiol 58(1): 35-38. et al. 2009).
ACTIVITY OF THE PHOSPHOESTERASE ENZYME IN STRAINS CM-Z6 AND CM-Z19
The phosphoesterase activity of the two strains are shown in Fig. 7. From the data in Fig. 7, it is clear that both strains have strong phosphoesterase activity during the growth process, and their phosphoesterase activity can be detected in their extracellular enzyme, intracellular enzyme and membrane-bound enzyme fractions. This may be the reason why both strains can effectively degrade a variety of organophosphorus pesticides. In addition, the phosphoesterase activity of strain CM-Z19 was significantly higher than that of strain CM-Z6 (P <0.05), which also confirms that the ability of strain CM-Z19 to degrade organophosphorus pesticides is better than that of strain CM-Z6. The experimental results also suggest that the phosphoesterase activity of these two strains is positively correlated with their degradation ability. However, because strain CM-Z6 could degrade TCP (the intermediate of CM) which is helpful for avoiding the accumulation of intermediate products and feedback inhibition and promoting the degradation reaction, it achieved faster degradation of CM than did strain CM-Z19.
Activity of the phosphoesterase enzyme (EE-extracellular enzyme, CE-crude enzyme, ME-membrane-bound enzyme, IE- intracellular enzyme).
CONCLUSION
In this study, strains CM-Z19 and CM-Z6, which were capable of highly degrading CM, were isolated from soil. Both strains had a high phosphoesterase activity and could degrade CM and a variety of organophosphorus pesticides. The phosphoesterase activity of the two strains was positively correlated with their degradation ability. Strain CM-Z19 had higher phosphoesterase activity and ability to degrade organophosphorus pesticide than did strain CM-Z6. However, strain CM-Z19 could not degrade TCP, and the accumulation of TCP in the degradation process was not conducive to further degradation of CM. Strain CM-Z6 could effectively degrade TCP and could degrade CM more quickly than could strain CM-Z19. Therefore, that rapid degradation of intermediates was considered to have an important effect on the degradation reaction.
ACKNOWLEGMENTS
This study was funded by the National Key Research and Development Plan of China (No. 2016YFD0600204) and China Postdoctoral Science Foundation (No. 2017M611827), and partially by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
REFERENCES
- ABBASSY MA, SALIM YMM, SHAWIR MS and NASSAR AMK. 2017. Disappearance and hazard quotient of chlorpyrifos-methyl, fipronil, and imidacloprid insecticides from dates. J Consum Prot Food S 12(3): 223-230.
- ABRAHAM J and SILAMBARASAN S. 2013. Biodegradation of chlorpyrifos and its hydrolyzing metabolite 3,5,6-trichloro-2-pyridinol by Sphingobacterium sp. JAS3. Process Biochem 48(10): 1559-1564.
- ABRAHAM J and SILAMBARASAN S. 2016. Biodegradation of chlorpyrifos and its hydrolysis product 3,5,6-trichloro-2-pyridinol using a novel bacterium Ochrobactrum sp. JAS2: a proposal of its metabolic pathway. Pestic Biochem Physiol 126(1): 13-21.
- ANWAR S, LIAQUAT F, KHAN QM, KHALID ZM and IQBAL S. 2009. Biodegradation of chlorpyrifos and its hydrolysis product 3,5,6-trichloro-2-pyridinol by Bacillus pumilus strain C2A1. J Hazard Mater 168(1): 400-405.
- BHADBHADE BJ, SARNAIK SS and KANEKAR PP. 2002. Biomineralization of an organophosphorus pesticide, monocrotophos, by soil bacteria. J Appl Microbiol 93(2): 224-234.
- BLANCHET P and ST-GEORGE A. 2010. Kinetics of chemical degradation of organophosphorus pesticides; hydrolysis of chlorpyrifos and chlorpyrifos-methyl in the presence of copper(ii). Pest Manag Sci 13(1): 85-91.
- CHO KM, MATH RK, ISLAM SM, LIM WJ, HONG SY, KIM JM, YUN MG,CHO JJ and YUN HD. 2009. Biodegradation of chlorpyrifos by lactic acid bacteria during kimchi fermentation. J Agric Food Chem 57(5): 1882-1889.
- CHOI WS and LEE SE. 2016. Toxicity of chlorpyrifos-methyl to Sitophilus zeamais collected in korea and biochemical differences. Entomol Res 46(1): 15-22.
- DAS S and SINGH DK. 2006. Purification and characterization of phosphotriesterases from Pseudomonas aeruginosa F10B and Clavibacter michiganense subsp. insidiosum SBL11. Can J Microbiol 52(2): 157-168.
- FINOCCHIARO R, MELI S and GENNARI M. 2004. Behavior of chlorpyrifos-methyl in soil and sediment. J Environ Sci Heal B 39(3): 381-392.
- JI X, WANG Q, ZHANG W and YIN F. 2016. Research advances in organophosphorus pesticide degradation: a review. Fresen Environ Bull 25(5): 1556-1561.
- KIM JR and AHN YJ. 2009. Identification and characterization of chlorpyrifos-methyl and 3,5,6-trichloro-2-pyridinol degrading Burkholderia sp. strain KR100. Biodegradation 20(4): 487-497.
- LAKSHMI CV, KUMAR M and KHANNA S. 2009. Biodegradation of chlorpyrifos in soil by enriched cultures. Curr Microbiol 58(1): 35-38.
- LEE SE. 2011. Purification and characterisation of a carboxylesterase from a chlorpyrifos-methyl-resistant strain of Oryzaephilus surinamensis (Coleoptera: Silvanidae). Aust J Entomol 50(2): 187-194.
- LI C, LIU H, HAO Z, KE H, TAO G and NI H. 2012. Characterization of a newly isolated highly effective 3,5,6-trichloro-2-pyridinol degrading strain Cupriavidus pauculus P2. Curr Microbiol 65(3): 231-236.
- MATTHEWS WA. 2010. The fate of chlorpyrifos-methyl in stored wheat: a comparison of a laboratory-scale experiment with a pilot-scale treatment. Pest Manag Sci 30(1): 21-29.
- MAYA K, SINGH RS, UPADHYAY SN and DUBEY SK. 2011. Kinetic analysis reveals bacterial efficacy for biodegradation of chlorpyrifos and its hydrolyzing metabolite TCP. Process Biochem 46(11): 2130-2136.
- MAYA K, UPADHYAY SN, SINGH RS and DUBEY SK. 2012. Degradation kinetics of chlorpyrifos and 3,5,6-trichloro-2-pyridinol (TCP) by fungal communities. Bioresour Technol 126: 216-223.
- MEIKLE RW and YOUNGSON CR. 1978. The hydrolysis rate of chlorpyrifos, o-o-diethyl o-(3,5,6-trichloro-2-pyridyl) phosphorothioate, and its dimethyl analog, chlorpyrifos-methyl, in dilute aqueous solution. Arch Environ Con Tox 7(1): 13-22.
- MOSCOSO F, TEIJIZ I, DEIVE FJ and SANROMÁN MA. 2013. Approaching chlorpyrifos bioelimination at bench scale bioreactor. Bioproc Biosyst Eng 36(9): 1303-1309.
- PENG G, LU Y, HE Q, MMEREKI D, ZHOU G, CHEN J and TANG X. 2016. Determination of 3,5,6-trichloro-2-pyridinol, phoxim and chlorpyrifos-methyl in water samples using a new pretreatment method coupled with high-performance liquid chromatography. J Sep Sci 38(24): 4204-4210.
- PRODHAN MDH, PAPADAKIS EN and PAPADOPOULOU-MOURKIDOU E. 2015. Determination of multiple pesticide residues in eggplant with liquid chromatography-mass spectrometry. Food Anal Method 8(1): 229-235.
- SAMBROOK J and RUSSELL DW. 1998. Molecular cloning: a laboratory manual. 3rd ed., Cold Spring Harbor Laboratory Press.
- SANGKHARAK K and PRASERTSAN P. 2012. Screening and identification of polyhydroxyalkanoates producing bacteria and biochemical characterization of their possible application. J Gen Appl Microbiol 58(3): 173-182.
- SHI XZ, GUO RJ, TAKAGI K, MIAO ZQ and LI SD. 2011. Chlorothalonil degradation by Ochrobactrum lupini strain TP-D1 and identification of its metabolites. World J Microb Biotech 27(8): 1755-1764.
- SILAMBARASAN S and ABRAHAM J. 2014. Efficacy of Ganoderma sp. JAS4 in bioremediation of chlorpyrifos and its hydrolyzing metabolite TCP from agricultural soil. J Basic Microb 54(1): 44-55.
- SONG X, ZHANG Z, LIU Y, ZHAN N and XIANG D. 2016. Isolation and identification of microorganisms causing mildew of fiberboard in Nanning,Guangxi. J Forest Eng 1(1): 78-82.
- SZPYRKA E, MATYASZEK A and SŁOWIK-BOROWIEC M. 2017. Dissipation of chlorantraniliprole, chlorpyrifos-methyl and indoxacarb-insecticides used to control codling moth (Cydia pomonella, L.) and leafrollers (Tortricidae) in apples for production of baby food. Environ Sci Pollut R 24(13): 12128-12135.
- TITUS KR. 1988. Degradation of chlorpyrifos and its hydrolysis product, 3,5,6-trichloro-2-pyridinol, in soil. J Environ Sci Heal B 23(6): 527-539.
- WANG S, ZHANG C and YAN Y. 2012. Biodegradation of methyl parathion and p-nitrophenol by a newly isolated Agrobacterium sp. strain Yw12. Biodegradation 23(1): 107-116.
- XIA M, HUAN C, JIANG T, ZHENG Z and OUYANG J. 2017. Molecular cloning and enzymological characteristics of a novel N-acetyl-β-D-glucosaminidase from Bacillus coagulans. J Forest Eng 2(2): 70-75.
- ZENG X. 2015. Identification of the pathogen causing Eucalyptus anthrax and laboratory screening of fungicides. China Forest Sci Technol (J Forest Eng) 29(4): 121-125.
- ZHU J, FU L, JIN C, MENG Z and YANG N. 2019. Study on the isolation of two atrazine-degrading bacteria and the development of a microbial agent. Microorganisms 7(3): 80.
- ZHU J, ZHAO Y, LI X, CHEN W and HUANG R. 2018b. Study on the characteristics and utilization of a triclopyr butoxyethyl ester degrading bacterium, Lactobacillus buchneri TBE-6. Fresen Environ Bull 27(5): 3156-3161.
- ZHU J, ZHAO Y, XIE K, LI X and CHEN Y. 2018a. Study on biodegradation of chlorpyrifos and utilization of a chlorpyrifos degrading bacterium, Bacillus amyloliquefaciens CZ-5. Fresen Environ Bull 27(2): 1196-1201.
Publication Dates
-
Publication in this collection
14 Oct 2019 -
Date of issue
2019
History
-
Received
11 July 2018 -
Accepted
26 Oct 2018