Abstracts
It is well established that genetic factors play an important role in the development of both type 2 diabetes mellitus (DM2) and obesity, and that genetically susceptible subjects can develop these metabolic diseases after being exposed to environmental risk factors. Therefore, great efforts have been made to identify genes associated with DM2 and/or obesity. Uncoupling protein 1 (UCP1) is mainly expressed in brown adipose tissue, and acts in thermogenesis, regulation of energy expenditure, and protection against oxidative stress. All these mechanisms are associated with the pathogenesis of DM2 and obesity. Hence, UCP1 is a candidate gene for the development of these disorders. Indeed, several studies have reported that polymorphisms -3826A/G, -1766A/G and -112A/C in the promoter region, Ala64Thr in exon 2 and Met299Leu in exon 5 of UCP1 gene are possibly associated with obesity and/or DM2. However, results are still controversial in different populations. Thus, the aim of this study was to review the role of UCP1 in the development of these metabolic diseases.
UCP1; obesity; type 2 diabetes mellitus; DNA polymorphisms; brown adipose tissue
Está bem estabelecido que fatores genéticos têm papel importante no desenvolvimento do diabetes melito tipo 2 (DM2) e obesidade e que indivíduos suscetíveis geneticamente podem desenvolver essas doenças metabólicas após exposição a fatores de risco ambientais. Assim, grandes esforços têm sido feitos para a identificação de genes associados ao DM2 e/ou à obesidade. A proteína desacopladora 1 (UCP1) é principalmente expressa no tecido adiposo marrom e atua na termogênese, regulação do gasto energético e proteção contra o estresse oxidativo, mecanismos associados tanto à patogênese do DM2 como à obesidade. Portanto, UCP1 é um gene candidato para o desenvolvimento dessas doenças. De fato, diversos estudos relataram que os polimorfismos -3826A/G, -1766A/G e -112A/C na região promotora, Ala64Thr no éxon 2 e Met299Leu no éxon 5 do gene UCP1 estão possivelmente associados à obesidade e/ou ao DM2. Entretanto, os resultados são ainda controversos em diferentes populações. Então, o objetivo deste estudo foi revisar o papel da UCP1 no desenvolvimento dessas doenças metabólicas.
UCP1; obesidade; diabetes melito tipo 2; polimorfismos de DNA; tecido adiposo marrom
REVIEW
The role of the uncoupling protein 1 (UCP1) on the development of obesity and type 2 diabetes mellitus
Papel da proteína desacopladora 1 (UCP1) no desenvolvimento da obesidade e do diabetes melito tipo 2
Letícia de Almeida Brondani; Taís Silveira Assmann; Guilherme Coutinho Kullmann Duarte; Jorge Luiz Gross; Luís Henrique Canani; Daisy Crispim
Endocrinology Division, Hospital de Clínicas de Porto Alegre, Universidade Federal do Rio Grande do Sul (HC-UFRGS), Porto Alegre, RS, Brazil
Correspondence to Correspondence to: Daisy Crispim Rua Ramiro Barcelos, 2350 prédio 12, 4° andar 90035-003 - Porto Alegre, RS, Brazil dcmoreira@hcpa.ufrgs.br
SUMMARY
It is well established that genetic factors play an important role in the development of both type 2 diabetes mellitus (DM2) and obesity, and that genetically susceptible subjects can develop these metabolic diseases after being exposed to environmental risk factors. Therefore, great efforts have been made to identify genes associated with DM2 and/or obesity. Uncoupling protein 1 (UCP1) is mainly expressed in brown adipose tissue, and acts in thermogenesis, regulation of energy expenditure, and protection against oxidative stress. All these mechanisms are associated with the pathogenesis of DM2 and obesity. Hence, UCP1 is a candidate gene for the development of these disorders. Indeed, several studies have reported that polymorphisms -3826A/G, -1766A/G and -112A/C in the promoter region, Ala64Thr in exon 2 and Met299Leu in exon 5 of UCP1 gene are possibly associated with obesity and/or DM2. However, results are still controversial in different populations. Thus, the aim of this study was to review the role of UCP1 in the development of these metabolic diseases. Arq Bras Endocrinol Metab. 2012;56(4):215-25
Keywords: UCP1; obesity; type 2 diabetes mellitus; DNA polymorphisms; brown adipose tissue
SUMÁRIO
Está bem estabelecido que fatores genéticos têm papel importante no desenvolvimento do diabetes melito tipo 2 (DM2) e obesidade e que indivíduos suscetíveis geneticamente podem desenvolver essas doenças metabólicas após exposição a fatores de risco ambientais. Assim, grandes esforços têm sido feitos para a identificação de genes associados ao DM2 e/ou à obesidade. A proteína desacopladora 1 (UCP1) é principalmente expressa no tecido adiposo marrom e atua na termogênese, regulação do gasto energético e proteção contra o estresse oxidativo, mecanismos associados tanto à patogênese do DM2 como à obesidade. Portanto, UCP1 é um gene candidato para o desenvolvimento dessas doenças. De fato, diversos estudos relataram que os polimorfismos -3826A/G, -1766A/G e -112A/C na região promotora, Ala64Thr no éxon 2 e Met299Leu no éxon 5 do gene UCP1 estão possivelmente associados à obesidade e/ou ao DM2. Entretanto, os resultados são ainda controversos em diferentes populações. Então, o objetivo deste estudo foi revisar o papel da UCP1 no desenvolvimento dessas doenças metabólicas. Arq Bras Endocrinol Metab. 2012;56(4):215-25
Descritores: UCP1; obesidade; diabetes melito tipo 2; polimorfismos de DNA; tecido adiposo marrom
INTRODUCTION
Type 2 diabetes mellitus (DM2) and obesity are common, multifactorial conditions for which susceptibility is determined by the joint actions of genetic and environmental factors (1). The prevalence of obesity and DM2 is increasing worldwide at an alarming rate, and both traits are associated with increased morbidity and mortality (2,3). The dramatic increase in the pre-valence of these disorders over the past two decades is mostly likely due to changes in diet and physical activity (4). However, it is believed that these environmental changes would only lead to DM2 and/or obesity under a permissible genetic background (1). Therefore, great efforts have been made to identify genes associa-ted with these disorders, and a number of studies have been focused on the genes related to energy expenditure, such as those encoding adrenergic receptors and mitochondrial uncoupling proteins (UCPs) (5).
Uncoupling protein 1 (UCP1) plays important roles in metabolic and energy balance and regulation, cold- and diet-induced thermogenesis, and in decreasing the production of reactive oxygen species (ROS) by mitochondria, which are mechanisms associated with the pathogenesis of obesity and/or DM2 (6,7). Thus, the aim of the present study was to review the role of UCP1 in relation to the development of these conditions.
Mitochondrial respiratory chain
Mitochondria are essential organelles in all eukaryotic cells and are involved in many processes that are crucial for cell survival and functioning, including energy production, redox control, calcium homeostasis, and certain metabolic and biosynthetic pathways. In addition, mitochondria are the main sources of ROS and often play a key role in physiological cell death mechanisms (7,8).
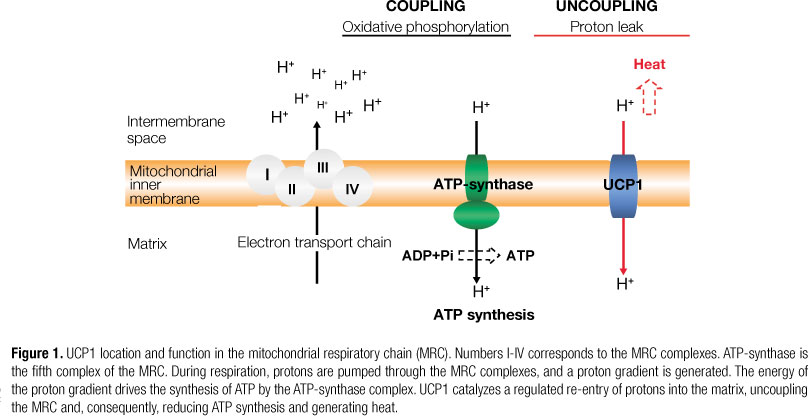
The main source of cell energy is the synthesis of ATP from ADP and inorganic phosphate (Pi) by oxidative phosphorylation (OXPHOS) carried out in the mitochondrial respiratory chain (MRC) (7). The MRC is located in the inner mitochondrial membrane, and is constituted by four multienzymatic complexes, an oligomeric protein complex (ATP-synthase), and two proteins responsible for electron transport, coenzyme Q (CoQ), and cytochrome c (Figure 1). OXPHOS involves the coupling of electron transport, by means of the complexes I-IV of the MRC, to the active pumping of protons across the inner mitochondrial membrane, and ATP formation by ATP-synthase (8).
Oxidation of reduced nutrient molecules by means of cellular metabolism yields electrons in the form of reduced hydrogen carriers (NADH and FADH2), which donate electrons to the MRC. The movement of electrons through the MRC is driven by a redox potential that is found across the chain. Complexes I, III, and IV pump protons across the inner membrane as electrons pass down the MRC. This produces an electrochemical potential difference across the inner membrane, known as proton-motive force, consisting mostly of an electrochemical gradient (membrane potential) and a chemical gradient (pH difference). The energy that is conserved in the proton gradient across the inner membrane is used by ATP-synthase to synthesize ATP as protons are transported back from the intermembrane space into the mitochondrial matrix. The final destination of the electrons is the generation of molecular oxygen, which is reduced to water by complex IV, in the last step of the MRC. Therefore, the process of substrate oxidation and oxygen reduction is also called respiration (8).
The coupling of respiration to ATP synthesis is not 100% efficient, and some of the energy is dissipated as heat. Partial uncoupling of respiration from ATP synthesis, also known as proton leak, can be mediated by UCPs and by other mitochondrial inner membrane proteins, such as adenine nucleotide translocase (ANT); which prevents the inhibition of MRC by excessive levels of ATP (6,9).
Although OXPHOS constitutes a vital part of cellular metabolism, the MRC is probably the most important site of ROS production (9). ROS correspond to a variety of molecules and free radicals (chemical species with one unpaired electron) derived from the metabolism of molecular oxygen. Superoxide anion (O2-) is the precursor of most ROS and a mediator in oxidative chain reactions (10). Dismutation of O2- (either spontaneously or by a reaction catalyzed by superoxide dismutases) produces hydrogen peroxide (H2O2) which, in turn, may be fully reduced to water or, in the presence of ferrous or cuprous ions, may form the highly reactive hydroxyl radical (+OH) (7,8). ROS normally exist in all aerobic cells in balance with biochemical antioxidants. Oxidative stress occurs when this critical balance is disrupted because of excess ROS, depletion of antioxidants, or both. This stress causes damage to cellular macromolecules, such as nucleic acids, proteins, lipids, and structural carbohydrates (11). Moreover, oxidative stress can also lead to cell death by necrosis or apoptosis, mechanisms involved in the pathogenesis of ageing and some disorders, such as DM2 and its chronic complications (11).
Uncoupling of the mitochondrial respiratory chain, changes in energy expenditure, and adaptive thermogenesis
Total body energy expenditure represents the conversion of oxygen and food (or storable forms of energy) to carbon dioxide, water, heat and "work" on the environment (10). Energy expenditure in humans can be subdivided into: 1) basal energy expenditure or resting metabolic rate (RMR), measured under resting conditions and required for normal cell functioning; 2) energy expenditure resulting from physical activity; and 3) energy expenditure attributed to adaptive thermogenesis (Figure 2) (7,10).
Uncoupling (proton leak) of the MRC constitutes a considerable part of the RMR (7). Approximately 20%-50% of total energy expenditure is due to proton leaks, with the skeletal muscle as the main contributor (10). Variations in the RMR are due to several determinants, including body composition (fat vs. fat-free mass), concentrations of steroid and thyroid hormones, genetic factors, and the activity of the sympathetic nervous system (7). It is known that low energy expenditure could predict future weight gain, and that only a slight imbalance between energy intake and energy expenditure is necessary for weight gain, if it persists for several years (10,11). Thus, increasing the energy expenditure by increasing proton leak in mitochondria has been recognized as an effective way to achieve weight loss (7).
Brown adipose tissue (BAT) is found in newborns, rodents and hibernating mammals, and is the main site of adaptive thermogenesis, which is defined as non-shivering heat production in response to environmental temperature or diet (10,12). As a result, thermogenesis in BAT has important roles in thermal and energetic balance and, when deficient, may lead to obesity (10). BAT is a metabolically active tissue, which consists of adipocytes rich in mitochondria and numerous small lipid droplets, and is heavily innervated by sympathetic nerves (7). This tissue differs from the white adipose tissue (WAT), which contains large lipid droplets and few mitochondria (7,12).
In fetuses and newborns, BAT has traditionally been regarded as occurring in specific depots, such as axillary, interscapular, perirenal, and periadrenal ones (10). At birth, human newborns have considerable amounts of BAT, corresponding to 1%-5% of total body weight. This amount takes care of heat generation for the body when the skeletal muscles are yet not able to make any controlled movements and thus, produce heat (10). Children have highly active functional BAT until 13-15 years of age; but, until a few years ago, it was thought that the quantity of BAT declined after puberty, and was rare in adults. Nevertheless, nowadays it is known that BAT can be found in adults in the presence of catecholamine-secreting tumors, such as pheochromocytomas and paragangliomas (13). Besides, some recent studies have shown that BAT in adults is highly active both functionally and metabolically, especially after chronic exposure to cold (14).
In 1978, Himms-Hagen and Desautels (15) showed that BAT metabolism played a role in the development of obesity, and that obese mice had a defect in the mechanisms necessary for the activation of BAT thermogenesis. After this pioneering work, many studies have also shown that defective BAT thermogenesis is involved in the development of obesity in most rodent models, and activation of BAT thermogenesis reduces weight gain in these animals (7,10). Studies in humans show that although the amount of BAT is reported to be decreased in healthy adults, it still responsible for 1%-2% of the energy expenditure, preventing weight gain of 1-2 kg a year (16,17). Interestingly, when heal-thy men that have BAT are exposed to cold (19°C), they have a 30% increase in energy expenditure compared to thermoneutrality (27°C), in contrast to those men with almost no BAT, who did not show any increa-se in cold-induced energy expenditure (18).
It has been suggested that development of ectopic BAT within the WAT may play an important role in preventing obesity (9). In agreement with this hypothesis, transgenic mice overexpressing UCP1 in their skeletal muscle or WAT develop a resistance to diet-induced obesity and DM2, and have a marked stimulation of fatty acid oxidation in muscles (7). In addition, Tiraby and cols. (19) reported that the adenovirus-mediated expression of human PGC-1α (PPARγ-coactivator-1α) increased the expression of UCP1, respiratory chain proteins, and fatty acid oxidation enzymes in human subcutaneous white adipocytes. Changes in the expression of other genes were also consistent with brown adipocyte mRNA expression profile. The authors concluded that human white adipocytes can therefore acquire typical features of brown fat cells following proper stimulation (19). These data indicate that moderate induction of UCP1 in WAT may be used to increase metabolic energy expenditure in obese subjects. Thus, specific uncoupling of adipocyte mitochondria remains an attractive target for the development of anti-obesity drugs (6,9).
Mitochondrial uncoupling proteins (UCPs)
UCPs 1, 2, 3, 4, and 5 are members of an anion-carrier protein family, and are located in the inner mitochondrial membrane (20). These proteins have similarities in their structures, but different tissue expression in mammals. The original UCP, UCP1, is mainly expressed in BAT (7). UCP2 is widely distributed, whereas UCP3 is mainly restricted to the skeletal muscle, and UCP4 and 5 are mainly expressed in the brain (7,8,20).
Over the last few years, several studies have shown that UCPs decrease metabolic efficiency by uncoupling substrate oxidation in mitochondria from ATP synthesis by the MRC. This is thought to be accomplished by promoting net translocation of protons from the intermembrane space, across the inner mitochondrial membrane, to the mitochondrial matrix, thereby dissipating the potential energy available for ATP synthesis, and consequently, decreasing ATP production (6,8). This uncoupling effect then leads to homologue- and tissue-specific functions, such as thermogenesis and energy expenditure (UCP1), regulation of free-fatty acids (FFAs) metabolism (UCP2 and UCP3), reduction in ROS formation (UCP1-3 and UCP5), and regulation of ATP-dependent processes (UCP2) (6,8,20).
Uncoupling protein 1 (UCP1)
Thermogenesis in BAT is due to UCP1, also called thermogenin or SLC25A7 (7). In 1985 the coding DNA sequence of UCP1 was cloned, and its amino acid sequence was determined (5). The UCP1 gene covers a 9kb region on chromosome 4 (region 4q28-q31), and contains 6 exons and 5 introns (Figure 3) (7). UCP1 is a 33-kDa dimeric protein that dissipates the pH gradient generated by OXPHOS (Figure 1), releasing chemical energy as heat (7,9). UCP1 gene expression is increased by cold, adrenergic stimulation, β3-agonists, retinoid and thyroid hormones, and cAMP (6,7,9). Its expression is activated by non-sterified fatty acids and inhibited by purine nucleotides (GDP, ATP and ADP) (6,20). Many studies based on the use of drugs that activate the β3-adrenergic receptor (β3-AR) confirmed that the sympathetic nervous system was the main trigger of UCP1 activation and induction (6). Moreover, uncoupling of the MRC by means of UCP1 action is only observed when cells are properly stimulated, for example, by norepinephrine (6,20). Norepinephrine stimulation of β3-AR results in three joint processes: 1) activation of p38 mitogen-activated protein kinase (MAPK) pathways that upregulate UCP1 synthesis; 2) activation of protein kinase A (PKA)-mediated path-ways that initiate lipolysis and release of acute regulators of UCP1, such as FFAs; and 3) inhibition of lysosomal pathways that degrade UCP1 (6). In addition, it is well known that PGC-1α plays a pivotal role in the regulation of UCP1 gene expression after adrenergic stimulation, by means of a MAPK pathway that is associated with the JNK-interacting protein (JIP) family of scaffold proteins (6,7).
The uncoupling activity of UCP1 is explained by its ability to transport protons across the inner mitochondrial membrane, in particular when FFAs bind to the protein. However, although CoQ has been described as a cofactor essential for its activity, the precise mechanism by which FFAs regulate transport through UCP1 is still a matter of intense debate (6,9). The main proposed mechanisms include the fatty acids protonophore (or flip-flop) model, and the channel (or proton buffering) model. In the flip-flop model, UCP1 is a carrier of fatty acid anions, which are transported by this protein from the matrix side to the intermembrane space. In this model, each fatty acid anion combines with a proton, becomes electrically neutral and flips back through the membrane, releasing the proton in the matrix (6). The channel model predicts a two-domain structure of UCP1 with a pore domain and a gating domain, which allows protons to pass through the UCP1. In this model, fatty acid carboxyl groups are involved in the proton transport by providing H+ buffering capacity (6). The arguments for and against each model were reviewed by Brand and cols. (21), in 1999.
Transgenic and knockout rodent models are common approaches to investigate the impact of overexpression or absence of a particular gene on the mouse phenotype. UCP1-knockout mice did not become obese, and merely showed an increased sensitivity to cold exposure (7). On the other hand, transgenic mice with increased UCP1 expression in WAT were obesity-resistant after being fed a diet rich in saturated fat (7). UCP1 has also been ectopically expressed in skeletal muscle of mice, and these animals showed improved glucose tolerance after being fed a high-fat diet, when compared with wild-type mice (7). In humans, UCP1 expression in the intraperitoneal fat of obese subjects is 50% lower than in normal weight subjects, in spite of the fact that the amount of BAT interspersed in WAT depots in adult individuals is relatively low (approximately 1 brown adipocyte/200 white adipocytes) (12,20). Until a few years ago, it was believed that UCP1 was expressed exclusively in BAT; but it has been recently reported that UCP1 mRNA expression and/or protein were also observed in WAT of mouse and humans, mammalian pancreatic islets, human skeletal muscle, bovine retina, human longitudinal smooth muscle layers, and rat and mouse thymocytes (22-26). However, the physiological role of UCP1 in these tissues is still a matter of debate (7). As already mentioned, uncoupling of the MRC due to UCP1 activity allows a more rapid flux of electrons through the inner mitochondrial membrane, reducing membrane potential and, consequently, decreasing ROS production (6,7,20). Therefore, the main role of UCP1 in these other tissues seems to be protection against oxidative stress (27,28). Superoxide anions could activate UCPs through lipid peroxidation products, such as 4-hydroxy-2-nonenal (HNE), which is a marker of oxidative stress and a direct activator of UCP1 (28,29). On the other hand, superoxide dismutase enzymes inhibit UCP1 activity (27). Interestingly, Cui and cols. (25) reported that endothelial cells from bovine retina incubated with high levels of glucose increased UCP1 and UCP2 expression, which protected them from ROS damage caused by glucotoxicity, suggesting a protective role of these UCPs in the pathogenesis of diabetic retinopathy, a chronic diabetic complication.
UCP1 gene polymorphisms associated with obesity and/or type 2 diabetes mellitus
Because UCP1 has been found to decrease membrane potential and increase energy expenditure, UCP1 gene is regarded as a candidate gene for obesity, DM2 or related-traits (7). For that reason, the relationship between UCP1 locus and susceptibility to these disorders has been investigated in a number of genetic studies, with particular attention being paid to -3826A/G, -1766A/G, and -112A/C polymorphisms in the promoter region; Ala64Thr polymorphism in exon 2; and Met299Leu polymorphism in exon 5 of the UCP1 gene (Figure 3). Results of these studies have been variable (Table 1): while some of them showed an association of one or more of these polymorphisms with obesity, DM2, body fat accumulation, body mass index (BMI), or other characteristics of metabolic syndrome (17,22,30-50), other studies were unable to find any association between these polymorphisms and these characteristics (51-61).
Most association studies about the effects of UCP1 gene polymorphisms focused on -3826A/G (rs1800592) polymorphism. The -3826G allele has been associated with reduced UCP1 mRNA expression in intraperitonial adipose tissue of obese subjects, indicating that this polymorphism has functional importance (62). Several independent studies support the association between the -3826G allele and obesity, BMI or other obesity-related parameters (35,36,40-43,46,63,64). Additionally, other studies indicate that the -3826G allele might be associated with reduced HDL-cholesterol levels (22,33,37,44), increased triglycerides (37,43), or LDL-cholesterol levels (39,44), and increased systolic and/or diastolic blood pressure (35,44).
Conversely, studies on the association of the effects of UCP1 gene polymorphisms on DM2 have shown controversial results: a few studies reported an association between the -3826G allele and DM2, insulin resistance (IR) or increased insulin or glucose levels (37,41), whereas other studies indicated that this polymorphism is not associated with these characteristics (32,36,40,52,54-56,59,60). A number of studies analyzed the association between other UCP1 gene polymorphisms and DM2. Mori and cols. (38) reported that the C allele of -112A/C polymorphism and Leu allele of Met299Leu polymorphism were associated with susceptibility to DM2 in Japanese subjects. A recent study showed that the -3826A/-112A/Met229 UCP1 haplotype was associated with increased risk for DM2 in Indian subjects (50). In addition, the study of Fukuyama and cols. (32) indicated that the -112A/C polymorphism was associated with both increased insulin resistance and hepatic lipid content in Japanese subjects with DM2.
Similar to UCP1, β3-AR is expressed in BAT and WAT and plays an important role in the induction of lipolysis and in the regulation of energy homeostasis (5). In addition, it is the main adrenoreceptor that stimulates UCP1 expression (6). The Trp64Arg polymorphism in the β3-AR gene has been associated with weight gain and other obesity-related indexes, as well as with insulin resistance in different populations [which were reviewed in (5)]. Interestingly, some studies have showed that a synergistic effect between the -3826A/G polymorphism (UCP1 gene) and the Trp64Arg polymorphism (β3-AR gene) is associated with an increased tendency for weight gain (46), lower RMR (58), resistance to weight loss (57), or subsequent weight-maintenance after a low-calorie diet (57). In contrast, other studies did not find any influence of the interaction between these two polymorphisms on the resistance to a low-calorie diet (65), BMI and triglyceride levels (40,43) or several metabolic parameters related to obesity and DM2 (60). Ethnical and age differences, as well as environmental factors and a synergistic effect with other genes might explain the controversial results among different investigations (5).
In brief, studies on these associations cited here indicated that the -3826A/G polymorphism contributes to the susceptibility to obesity. On the other hand, results reported by other studies on the effects of -3826A/G polymorphism and other UCP1 gene polymorphisms on lipid profile, blood pressure or DM2 are still inconclusive.
CONCLUSION
Several studies have contributed to the understanding of the mechanisms underlying BAT function and UCP1 activity in this tissue. Interestingly, recent studies have shown that UCP1 can also be detected in pancreatic islets, WAT, skeletal muscle, longitudinal smooth muscle layers, retina, and thymus. However, the physiological functions of UCP1 in these tissues are not as well established as in BAT, and future studies will determine the role of UCP1 in these tissues.
Obesity and DM2 are multifactorial diseases associated with both genetic and environmental factors. Knowledge on factors associated with these disorders will allow us to better understand them, and may provide us with more effective approaches to treatment and prevention. UCP1 plays important roles in thermogenesis, regulation of energy expenditure, and in decreasing oxidative stress, which are mechanisms associated with the pathogenesis of obesity and DM2. Thus, UCP1 gene is an excellent candidate for these disorders. Indeed, numerous studies strongly suggest that the UCP1 -3826A/G polymorphism is associated with obesity. Further studies are required to investigate UCP1 gene polymorphism in different populations in order to confirm the association between these polymorphisms and DM2, and also to elucidate the molecular mechanisms of association between UCP1 polymorphisms and obesity, DM2, and related-phenotypes.
Acknowledgments: this study was partially supported by grants from Conselho Nacional de Desenvolvimento Científico e Tecnólogico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Capes) and Fundo de Incentivo à Pesquisa e Eventos (Fife) of Hospital de Clínicas de Porto Alegre.
Disclosure: no potential conflict of interest relevant to this article was reported.
Received on May/1/2012
Accepted on June/10/2012
- 1. Vimaleswaran KS, Loos RJ. Progress in the genetics of common obesity and type 2 diabetes. Expert Rev Mol Med. 2010;12:e7.
- 2. Flegal KM, Graubard BI, Williamson DF, Gail MH. Excess deaths associated with underweight, overweight, and obesity. JAMA. 2005;293(15):1861-7. Epub 2005/4/21.
- 3. Narayan KM, Boyle JP, Thompson TJ, Sorensen SW, Williamson DF. Lifetime risk for diabetes mellitus in the United States. JAMA. 2003;290(14):1884-90.
- 4. Hill JO, Wyatt HR, Reed GW, Peters JC. Obesity and the environment: where do we go from here? Science. 2003;299(5608):853-5.
- 5. Jia JJ, Tian YB, Cao ZH, Tao LL, Zhang X, Gao SZ, et al. The polymorphisms of UCP1 genes associated with fat metabolism, obesity and diabetes. Mol Biol Rep. 2010;37(3):1513-22.
- 6. Azzu V, Brand MD. The on-off switches of the mitochondrial uncoupling proteins. Trends Biochem Sci. 2010;35(5):298-307.
- 7. Dalgaard LT, Pedersen O. Uncoupling proteins: functional characteristics and role in the pathogenesis of obesity and Type II diabetes. Diabetologia. 2001;44(8):946-65.
- 8. Souza BM, Assmann TS, Kliemann LM, Gross JL, Canani LH, Crispim D. The role of uncoupling protein 2 (UCP2) on the development of type 2 diabetes mellitus and its chronic complications. Arq Bras Endocrinol Metabol. 2011;55(4):239-48.
- 9. Ricquier D. Respiration uncoupling and metabolism in the control of energy expenditure. Proc Nutr Soc. 2005;64(1):47-52.
- 10. Lowell BB, Spiegelman BM. Towards a molecular understanding of adaptive thermogenesis. Nature. 2000;404(6778):652-60.
- 11. Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307(5708):384-7.
- 12. Virtanen KA, Nuutila P. Brown adipose tissue in humans. Curr Opin Lipidol. 2011;22(1):49-54.
- 13. Fukuchi K, Tatsumi M, Ishida Y, Oku N, Hatazawa J, Wahl RL. Radionuclide imaging metabolic activity of brown adipose tissue in a patient with pheochromocytoma. Exp Clin Endocrinol Diabetes. 2004;112(10):601-3.
- 14. Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360(15):1518-25.
- 15. Himms-Hagen J, Desautels M. A mitochondrial defect in brown adipose tissue of the obese (ob/ob) mouse: reduced binding of purine nucleotides and a failure to respond to cold by an increase in binding. Biochem Biophys Res Commun. 1978;83(2):628-34.
- 16. Bouillaud F, Villarroya F, Hentz E, Raimbault S, Cassard AM, Ricquier D. Detection of brown adipose tissue uncoupling protein mRNA in adult patients by a human genomic probe. Clin Sci (Lond). 1988;75(1):21-7.
- 17. Hamann A, Tafel J, Büsing B, Münzberg H, Hinney A, Mayer H, et al. Analysis of the uncoupling protein-1 (UCP1) gene in obese and lean subjects: identification of four amino acid variants. Int J Obes Relat Metab Disord. 1998;22(9):939-41.
- 18. Yoneshiro T, Aita S, Matsushita M, Kameya T, Nakada K, Kawai Y, et al. Brown adipose tissue, whole-body energy expenditure, and thermogenesis in healthy adult men. Obesity (Silver Spring). 2011;19(1):13-6.
- 19. Tiraby C, Tavernier G, Lefort C, Larrouy D, Bouillaud F, Ricquier D, et al. Acquirement of brown fat cell features by human white adipocytes. J Biol Chem. 2003;278(35):33370-6.
- 20. Fisler JS, Warden CH. Uncoupling proteins, dietary fat and the metabolic syndrome. Nutr Metab (Lond). 2006;3:38.
- 21. Brand MD, Brindle KM, Buckingham JA, Harper JA, Rolfe DF, Stuart JA. The significance and mechanism of mitochondrial proton conductance. Int J Obes Relat Metab Disord. 1999;23 Suppl 6:S4-11.
- 22. Sale MM, Hsu FC, Palmer ND, Gordon CJ, Keene KL, Borgerink HM, et al. The uncoupling protein 1 gene, UCP1, is expressed in mammalian islet cells and associated with acute insulin response to glucose in African American families from the IRAS Family Study. BMC Endocr Disord. 2007;7:1.
- 23. Nagase I, Yoshida T, Kumamoto K, Umekawa T, Sakane N, Nikami H, et al. Expression of uncoupling protein in skeletal muscle and white fat of obese mice treated with thermogenic beta 3-adrenergic agonist. J Clin Invest. 1996;97(12):2898-904.
- 24. Nibbelink M, Moulin K, Arnaud E, Duval C, Pénicaud L, Casteilla L. Brown fat UCP1 is specifically expressed in uterine longitudinal smooth muscle cells. J Biol Chem. 2001;276(50):47291-5.
- 25. Cui Y, Xu X, Bi H, Zhu Q, Wu J, Xia X, et al. Expression modification of uncoupling proteins and MnSOD in retinal endothelial cells and pericytes induced by high glucose: the role of reactive oxygen species in diabetic retinopathy. Exp Eye Res. 2006;83(4):807-16.
- 26. Carroll AM, Haines LR, Pearson TW, Fallon PG, Walsh CM, Brennan CM, et al. Identification of a functioning mitochondrial uncoupling protein 1 in thymus. J Biol Chem. 2005;280(16):15534-43.
- 27. Echtay KS, Roussel D, St-Pierre J, Jekabsons MB, Cadenas S, Stuart JA, et al. Superoxide activates mitochondrial uncoupling proteins. Nature. 2002;415(6867):96-9.
- 28. Brand MD, Affourtit C, Esteves TC, Green K, Lambert AJ, Miwa S, et al. Mitochondrial superoxide: production, biological effects, and activation of uncoupling proteins. Free Radic Biol Med. 2004;37(6):755-67.
- 29. Echtay KS, Esteves TC, Pakay JL, Jekabsons MB, Lambert AJ, Portero-Otín M, et al. A signalling role for 4-hydroxy-2-nonenal in regulation of mitochondrial uncoupling. EMBO J. 2003;22(16):4103-10.
- 30. Herrmann SM, Wang JG, Staessen JA, Kertmen E, Schmidt-Petersen K, Zidek W, et al. Uncoupling protein 1 and 3 polymorphisms are associated with waist-to-hip ratio. J Mol Med. 2003;81(5):327-32.
- 31. Kim KS, Cho DY, Kim YJ, Choi SM, Kim JY, Shin SU, et al. The finding of new genetic polymorphism of UCP-1 A-1766G and its effects on body fat accumulation. Biochim Biophys Acta. 2005;1741(1-2):149-55.
- 32. Fukuyama K, Ohara T, Hirota Y, Maeda K, Kuno S, Zenibayashi M, et al. Association of the -112A>C polymorphism of the uncoupling protein 1 gene with insulin resistance in Japanese individuals with type 2 diabetes. Biochem Biophys Res Commun. 2006;339(4):1212-6.
- 33. Hamada T, Kotani K, Nagai N, Tsuzaki K, Matsuoka Y, Sano Y, et al. Low-calorie diet-induced reduction in serum HDL cholesterol is ameliorated in obese women with the -3826 G allele in the uncoupling protein-1 gene. Tohoku J Exp Med. 2009;219(4):337-42.
- 34. Sramkova D, Krejbichova S, Vcelak J, Vankova M, Samalikova P, Hill M, et al. The UCP1 gene polymorphism A-3826G in relation to DM2 and body composition in Czech population. Exp Clin Endocrinol Diabetes. 2007;115(5):303-7.
- 35. Forga L, Corbalán M, Marti A, Fuentes C, Martínez-González MA, Martínez A. [Influence of the polymorphism 03826 A > G in the UCP1 gene on the components of metabolic syndrome]. An Sist Sanit Navar. 2003;26(2):231-6.
- 36. Ramis JM, González-Sánchez JL, Proenza AM, Martínez-Larrad MT, Fernández-Pérez C, Palou A, et al. The Arg64 allele of the beta 3-adrenoceptor gene but not the -3826G allele of the uncoupling protein 1 gene is associated with increased leptin levels in the Spanish population. Metabolism. 2004;53(11):1411-6.
- 37. KieĆ-Wilk B, WybraŃska I, Malczewska-Malec M, LeszczyŃska-GoŁabek L, Partyka L, NiedbaŁ S, et al. Correlation of the -3826A >G polymorphism in the promoter of the uncoupling protein 1 gene with obesity and metabolic disorders in obese families from southern Poland. J Physiol Pharmacol. 2002;53(3):477-90.
- 38. Mori H, Okazawa H, Iwamoto K, Maeda E, Hashiramoto M, Kasuga M. A polymorphism in the 5' untranslated region and a Met229>Leu variant in exon 5 of the human UCP1 gene are associated with susceptibility to type II diabetes mellitus. Diabetologia. 2001;44(3):373-6.
- 39. Proenza AM, Poissonnet CM, Ozata M, Ozen S, Guran S, Palou A, et al. Association of sets of alleles of genes encoding beta3-adrenoreceptor, uncoupling protein 1 and lipoprotein lipase with increased risk of metabolic complications in obesity. Int J Obes Relat Metab Disord. 2000;24(1):93-100.
- 40. Hayakawa T, Nagai Y, Taniguchi M, Yamashita H, Takamura T, Abe T, et al. Phenotypic characterization of the beta3-adrenergic receptor mutation and the uncoupling protein 1 polymorphism in Japanese men. Metabolism. 1999;48(5):636-40.
- 41. Heilbronn LK, Kind KL, Pancewicz E, Morris AM, Noakes M, Clifton PM. Association of -3826 G variant in uncoupling protein-1 with increased BMI in overweight Australian women. Diabetologia. 2000;43(2):242-4.
- 42. Nakano T, Shinka T, Sei M, Sato Y, Umeno M, Sakamoto K, et al. A/G heterozygote of the A-3826G polymorphism in the UCP-1 gene has higher BMI than A/A and G/G homozygote in young Japanese males. J Med Invest. 2006;53(3-4):218-22.
- 43. Matsushita H, Kurabayashi T, Tomita M, Kato N, Tanaka K. Effects of uncoupling protein 1 and beta3-adrenergic receptor gene polymorphisms on body size and serum lipid concentrations in Japanese women. Maturitas. 2003;45(1):39-45.
- 44. Oh HH, Kim KS, Choi SM, Yang HS, Yoon Y. The effects of uncoupling protein-1 genotype on lipoprotein cholesterol level in Korean obese subjects. Metabolism. 2004;53(8):1054-9.
- 45. Shin HD, Kim KS, Cha MH, Yoon Y. The effects of UCP-1 polymorphisms on obesity phenotypes among Korean female subjects. Biochem Biophys Res Commun. 2005;335(2):624-30.
- 46. Clément K, Ruiz J, Cassard-Doulcier AM, Bouillaud F, Ricquier D, Basdevant A, et al. Additive effect of A>G (-3826) variant of the uncoupling protein gene and the Trp64Arg mutation of the beta 3-adrenergic receptor gene on weight gain in morbid obesity. Int J Obes Relat Metab Disord. 1996;20(12):1062-6.
- 47. Franco-Hincapié L, Duque CE, Parra MV, Gallego N, Villegas A, Ruiz-Linares A, et al. [Association between polymorphism in uncoupling proteins and type 2 diabetes in a northwestern Colombian population]. Biomedica. 2009;29(1):108-18.
- 48. Kim JY, Lee SS. The effects of uncoupling protein 1 and beta3-adrenergic receptor gene polymorphisms on weight loss and lipid profiles in obese women. Int J Vitam Nutr Res. 2010;80(2):87-96.
- 49. Cha MH, Kang BK, Suh D, Kim KS, Yang Y, Yoon Y. Association of UCP1 genetic polymorphisms with blood pressure among Korean female subjects. J Korean Med Sci. 2008;23(5):776-80.
- 50. Vimaleswaran KS, Radha V, Ghosh S, Majumder PP, Rao MR, Mohan V. A haplotype at the UCP1 gene locus contributes to genetic risk for type 2 diabetes in Asian Indians (CURES-72). Metab Syndr Relat Disord. 2010;8(1):63-8.
- 51. Shen ZN, Wang XS, Bai H, Fan P, Liu R, Liu Y, et al. [Analysis of -3826A/G polymorphism in the promoter of the uncoupling protein-1 gene in Chinese non-obese and obese populations]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2009;26(5):555-61.
- 52. Mottagui-Tabar S, Hoffstedt J, Brookes AJ, Jiao H, Arner P, Dahlman I. Association of ADRB1 and UCP3 gene polymorphisms with insulin sensitivity but not obesity. Horm Res. 2008;69(1):31-6.
- 53. Nieters A, Becker N, Linseisen J. Polymorphisms in candidate obesity genes and their interaction with dietary intake of n-6 polyunsaturated fatty acids affect obesity risk in a sub-sample of the EPIC-Heidelberg cohort. Eur J Nutr. 2002;41(5):210-21.
- 54. Urhammer SA, Hansen T, Borch-Johnsen K, Pedersen O. Studies of the synergistic effect of the Trp/Arg64 polymorphism of the beta3-adrenergic receptor gene and the -3826 A>G variant of the uncoupling protein-1 gene on features of obesity and insulin resistance in a population-based sample of 379 young Danish subjects. J Clin Endocrinol Metab. 2000;85(9):3151-4.
- 55. Sivenius K, Valve R, Lindi V, Niskanen L, Laakso M, Uusitupa M. Synergistic effect of polymorphisms in uncoupling protein 1 and beta3-adrenergic receptor genes on long-term body weight change in Finnish type 2 diabetic and non-diabetic control subjects. Int J Obes Relat Metab Disord. 2000;24(4):514-9.
- 56. Boullu-Sanchis S, Leprêtre F, Hedelin G, Donnet JP, Schaffer P, Froguel P, et al. Type 2 diabetes mellitus: association study of five candidate genes in an Indian population of Guadeloupe, genetic contribution of FABP2 polymorphism. Diabetes Metab. 1999;25(2):150-6.
- 57. Fogelholm M, Valve R, Kukkonen-Harjula K, Nenonen A, Hakkarainen V, Laakso M, et al. Additive effects of the mutations in the beta3-adrenergic receptor and uncoupling protein-1 genes on weight loss and weight maintenance in Finnish women. J Clin Endocrinol Metab. 1998;83(12):4246-50.
- 58. Valve R, Heikkinen S, Rissanen A, Laakso M, Uusitupa M. Synergistic effect of polymorphisms in uncoupling protein 1 and beta3-adrenergic receptor genes on basal metabolic rate in obese Finns. Diabetologia. 1998;41(3):357-61.
- 59. Evans D, Minouchehr S, Hagemann G, Mann WA, Wendt D, Wolf A, et al. Frequency of and interaction between polymorphisms in the beta3-adrenergic receptor and in uncoupling proteins 1 and 2 and obesity in Germans. Int J Obes Relat Metab Disord. 2000;24(10):1239-45.
- 60. Schäffler A, Palitzsch KD, Watzlawek E, Drobnik W, Schwer H, Schölmerich J, et al. Frequency and significance of the A>G (-3826) polymorphism in the promoter of the gene for uncoupling protein-1 with regard to metabolic parameters and adipocyte transcription factor binding in a large population-based Caucasian cohort. Eur J Clin Invest. 1999;29(9):770-9.
- 61. Gagnon J, Lago F, Chagnon YC, Pérusse L, Näslund I, Lissner L, et al. DNA polymorphism in the uncoupling protein 1 (UCP1) gene has no effect on obesity related phenotypes in the Swedish Obese Subjects cohorts. Int J Obes Relat Metab Disord. 1998;22(6):500-5.
- 62. Esterbauer H, Oberkofler H, Liu YM, Breban D, Hell E, Krempler F, et al. Uncoupling protein-1 mRNA expression in obese human subjects: the role of sequence variations at the uncoupling protein-1 gene locus. J Lipid Res. 1998;39(4):834-44.
- 63. Sramkova D, Krejbichova S, Vcelak J, Vankova M, Samalikova P, Hill M, et al. The UCP1 gene polymorphism A-3826G in relation to DM2 and body composition in Czech population. Exp Clin Endocrinol Diabetes. 2007;115(5):303-7. Epub 2007/05/23.
- 64. Cha MH, Kim KS, Suh D, Chung SI, Yoon Y. A UCP1-412A>C polymorphism is associated with abdominal fat area in Korean women. Hereditas. 2008;145(5):231-7.
- 65. Fumeron F, Durack-Bown I, Betoulle D, Cassard-Doulcier AM, Tuzet S, Bouillaud F, et al. Polymorphisms of uncoupling protein (UCP) and beta 3 adrenoreceptor genes in obese people submitted to a low calorie diet. Int J Obes Relat Metab Disord. 1996;20(12):1051-4.
Publication Dates
-
Publication in this collection
11 July 2012 -
Date of issue
June 2012
History
-
Received
01 May 2012 -
Accepted
10 June 2012