ABSTRACT
BACKGROUND:
Colorectal cancer (CRC) is one of the leading causes of cancer worldwide. Early diagnostic methods using serum biomarkers are required. The study of omics, most recently lipidomics, has the purpose of analyzing lipids for a better understanding of human lipidoma. The evolution of mass spectrometry methods, such as MALDI-MS technology, has enabled the detection and identification of a wide variety of lipids with great potential to open new avenues for predictive and preventive medicine.
OBJECTIVE:
To determine the lipid profile of patients with colorectal cancer and polyps.
METHODS:
Patients with stage I-III CRC, adenomatous polyps and individuals with normal colonoscopy were selected. All patients underwent peripheral blood collection for lipid extraction. The samples were analyzed by MALDI-MS technique for lipid identification.
STATISTICAL ANALYSIS:
Univariate and multivariate (principal component analysis [PCA] and discriminant analysis by partial least squares [PLS-DA]) analyses workflows were applied to the dataset, using MetaboAnalyst 3.0 software. The ions were identified according to the class of lipids using the online database Lipid Maps (http://www.lipidmaps.org).
RESULTS:
We included 88 individuals, 40 with CRC, 12 with polyps and 32 controls. Boxplot analysis showed eight VIP ions in the three groups. Differences were observed between the cancer and control groups, as well as between cancer and polyp, but not between polyps and control. The polyketide (810.1) was the lipid represented in cancer and overrepresented in polyp and control. Among the patients with CRC we observed differences between lipids with lymph node invasion (N1-2) compared to those without lymph node invasion (N).
CONCLUSION:
Possible lipid biomarkers were identified among cancer patients compared to control and polyp groups. The polyketide lipid (810.1) was the best biomarker to differentiate the cancer group from control and polyp. We found no difference between the biomarkers in the polyp group in relation to the control.
HEADINGS:
Colorectal neoplasms; Biomarkers; Mass spectrometry; Lipid metabolism
RESUMO
CONTEXTO:
O câncer colorretal (CCR) é, mundialmente, uma das principais causas de câncer. Métodos de diagnóstico precoce através de biomarcadores séricos são necessários. O estudo das ômicas, mais recentemente a lipidômica, tem a finalidade de analisar os lipídeos para melhor compreensão do lipidoma humano. A evolução dos métodos de espectrometria de massa, como a tecnologia por MALDI-MS, possibilitou a detecção e a identificação de uma ampla variedade de lipídeos com grande potencial para abrir novos caminhos para a medicina preditiva e preventiva.
OBJETIVO:
Determinar o perfil lipidômico de pacientes com câncer colorretal e pólipos.
MÉTODOS:
Foram selecionados pacientes com CCR estádio I-III, com pólipos adenomatosos e indivíduos com colonoscopia normal. Todos os pacientes foram submetidos a coleta do sangue periférico para extração do lipídeo. As amostras foram analisadas por técnica de MALDI-MS para a identificação dos lipídeos.
ANÁLISE ESTATÍSTICA:
Para análise univariada e multivariada foram utilizados a análise de componentes principais (PCA) e a análise discriminante pelos quadrados mínimos (PLS-DA). Os íons foram identificados de acordo com a classe de lipídeos usando-se o Lipid Maps (http://www.lipidmaps.org).
RESULTADOS:
Foram incluídos 88 indivíduos, 40 com CCR, 12 com pólipos e 32 controles. A análise de boxbolt evidenciou oito íons VIP nos três grupos. Observou-se diferenças entre os grupos câncer e controle, assim como entre câncer e pólipo, mas não entre pólipos e controle. O policetídeo (810,1) foi o lipídeo hipo-representado no câncer e hiperrepresentado no pólipo e controle. Entre os pacientes com CCR observamos diferenças entre os lipídeos com invasão linfonodal (N1-2) comparados aos sem invasão linfonodal (N0).
CONCLUSÃO:
Foram identificados possíveis biomarcadores lipídicos entre os pacientes com câncer comparados aos grupos controle e pólipo. O lipídeo policetídeo (810,1) foi o melhor biomarcador para diferenciar o grupo câncer do controle e pólipo. Não encontramos diferença entre os biomarcadores no grupo pólipo em relação ao controle.
DESCRITORES:
Neoplasias colorretais; Biomarcadores; Espectrometria de massas; Metabolismo dos lipídeos
INTRODUCTION
Colorectal cancer (CRC) is worldwide, the third cause of cancer and the second cause of death. An estimated 1.8 million new cases and 881,000 deaths occurred by this cancer yearly11. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortalty worldwide for 36 cancers in 185 countries CA Cancer J Clin. 2018;68:394-424.. According to the Brazilian statistics, CRC is the second cause of cancer for women and the second for men22. National Cancer Institute (INCA). Estimate 2018: Cancer incidence in Brazil. [Internet]. [Acessed 2019 June 19]. Available from: https://www.inca.gov.br/search/conteudo/estimativa
https://www.inca.gov.br/search/conteudo/...
.
CRC screening is necessary to detect polyps and cancer precociously33. Dumoulin FL Hildenbrand R. Endoscopic resection techniques for colorectal neoplasia: current developments. World J Gastroenterol. 2019;25:300-7.. The diagnosis at initial stages is crucial for acquiring the best prognosis. Colonoscopy, sigmoidoscopy and the fecal occult blood test (FOBT) are the main methods for CRC screening. Colonoscopy is a highly sensitive exam for identifying adenomas and cancers but is an invasive and expensive method with low adhesion even in developed countries44. Issa IA, Noureddine M. Colorectal cancer screening: an updated review of the available options. World J Gastroenterol . 2017;23:5086-96.. The FOBT has low sensitivity and specificity, particularly for adenomas, but is a non-invasive and low-cost exam that reduces CRC mortality up to 16%. Sigmoidoscopy, allow partial visualization of the colon but also reduced mortality up to 30%55. Bray C, Bell LN, Liang H, Collins D, Yale SH. Colorectal Cancer Screening. WMJ. 2017;116:27-33..
There is an urgent need to develop simple tests, less invasive, with high sensitivity and specificity. In recent decades, many studies have been conducted to detect molecular markers in blood, stools and urine. Mutations or methylations alterations on DNA may be interesting ways for CRC diagnosis66. Yiu AJ, Yiu CY. Biomarkers in Colorectal. Anticancer Res. 2016;36:1093-102..
The MALDI-TOF MS technique is a high sensitivity and specificity analytical test for the diagnosis of cancer, applied to identify a multitude of cancer types such as gastrointestinal, lung, renal, bladder, ovarian, leukemia, melanoma and breast carcinoma. The analysis of lipid profiles can significantly contribute to the indication of some pathologies such as cancer77. Wang C, Miao Wang M, Han X. Applications of Mass Spectrometry for Cellular Lipid Analysis. Mol Biosyst. 2015;11:698-713.. Qualitative and quantitative assessment of lipids in blood and other body fluids may reveal new biomarkers for early diagnosis of cancer, as well as to follow cancer patients after treatment88. Li M, Yang L, Bai Y, Liu H. Analytical Methods in Lipidomics and Their Applications. Anal Chem. 2014;86:161-75..
The family of lipid is composed of a wide variety of molecular species, divided into eight classes that include 1,680,000 species complex lipids. These are involved in various physiological processes regulated by thousands of metabolic pathways. Changes in expression levels of individual lipid species may be associated with numerous diseases including cancer99. Ingólfsson HI, Melo MN, van Eerden FJ, Arnarez C, Lopez CA, Wassenaar TA, et al. Lipid organization of the plasma membrane. J Am Chem Soc. 2014;136:14554-9..
There are two strategies for lipid analysis. The first one that focus on known lipids and the other one that analysis non-target lipid that provides a vast amount of data, which require the assistance of bioinformatics for data processing. This process includes three phases: the first refers to the principal component analysis (PCA) and discriminatory analysis of partial least squares (PLS-DA) that aims to identify the differential lipids. The second strategy has the purpose of recovery of the database coupled with MS/MS spectra for identification of different lipids. The third and final phase targeted the interpretation of the data for the acquisition of meaningful information1010. Li L, Han J, Wang Z, Liu J, Wei J, Xiong S, et al. Mass spectrometry methodology in lipid analysis. Int J Mol Sci, 2014;15:1492-507..
The mass spectrometry (MS) is a single analytical method that allows the identification of a wide range of molecules along the surface of a biological sample. With the advances in MS technologies, many analytical equipment was developed allowing different ionization and fragmentation approaches. The MALDI technique is the most currently used as a high-performance tool, due to its high sensitivity. MALDI-TOF/MS has become a promising tool between the present analytical platforms for lipidomic research, proving to be an attractive platform for application in lipidomics for pre-clinical biomarker studies1111. Calvano CD, van der Werf ID, Palmisano F, Sabbatini L. Identification of lipid- and protein-based binders in paintings by direct on-plate wet chemistry and matrix-assisted laser desorption ionization mass spectometry. Anal Bioanal Chem. 2015;407:1015-22.,1212. Takats Z, Strittmatter N, McKenzie JS. Ambient Mass Spectrometry in Cancer Research. Adv Cancer Res. 2017;134:231-56..
The objective of the study was to compare the plasmatic lipid profiles of patients with colorectal cancer to controls and patients with polyps.
METHODS
Patients were selected sequentially from the Clinical Oncology, Surgical Gastroenterology and Endoscopy Division of the Federal University of Sao Paulo. The study enrolled patients with colorectal cancer or polyps and patients with normal colonoscopy of both genders, aged between 20 and 80 years.
Colon cancer patients before surgical resection of the tumor and rectal cancer patients before neoadjuvant treatment were included and compose the cancer group. Patients undergoing colonoscopy with normal colonoscopy or with adenomas polyps were included before polypectomy and compose the control and the polyps’ group respectively. Patients with inflammatory bowel disease, familial adenomatous polyposis or previous history of cancer were excluded.
The study was approved by the Ethics and Clinical Research Committee Coordenadoria de Ensino e Pesquisa do Hospital São Paulo - HU/UNIFESP under no. 72523. All the patients were informed about the study and signed an Informed Consent term prior to their inclusion.
All patients underwent collection of 10 mL of peripheral blood in tubes with EDTA. After centrifugation, the plasma was stored at -80°C for further lipids analysis.
Lipid extraction
Aliquots were thawed at room temperature for lipids extraction following the protocol described by Bligh-Dyer (1959)1313. Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1957;37:911-7.. In a tube 300 μL of the sample were added to 375 μL of chloroform and 750 μL of methanol. After homogenization, 187.5 μL of chloroform and 150 μL of distilled water were added to supernatant and centrifuged at 3000 x g for 1 minute. Approximately 200 μL of the organic phase containing the lipids were gently transferred to a new tube, which remained open at room temperature for solvent evaporation.
Samples were resuspended in 8 μL methanol, placed in duplicate spot of 1 μL by the MTP adapter 384 (Bruker Daltonics, Bremen, Germany) and coated with 1 μL of 2,5-dihydroxybenzoic acid (DHB) and alfacyano-4-hydroxycinnamic (CHCA) both in the concentration of 7 mg/mL trifluoroacetic acid (TFA) 0.01% dissolved in 70% methanol and 30% water. The acquisition of the spectra was performed in the AUTOFLEX Speedy TOF/TOF (Bruker Daltonics, Bremen, Germany) in positive mode at a range of 700-1200 m/z with the frequency 500 water 1000 shots/sec and power 90%.
Statistical analysis
A list of ions and their intensities had been generated by Flexanalysis software (Bruker Daltonics, Bremen, Germany) and afterwards exported to an Excell spreadsheet. The statistical processing was performed in MetaboAnalyst software 2.0 (http://www.metaboanalyst.ca). The principal component analysis (PCA) and discriminant analysis by least squares (PLS-DA) were applied to evaluate the quality of the models. To find different changes on circulating lipids between the groups, 15 VIP lipids (variable importance in discriminant analysis) was obtained.
The ROC curves were generated, the area under the curve was generated to evaluate the cutoff, sensitivity, specificity for each VIP lipid in the groups.
The ions were identified by lipid subclasses by Simlipid 4 software (Premier Biosoft, Califonia, USA) and online database Lipid Maps (National Institute of Medical Sciences of the United States) (http://www.lipidmaps.org). They were identified and accepted hydrogen adducts (M+), sodium (Na+) and potassium (K+), since they form part of the composition of solvents.
RESULTS
Eighty-eight patients were included, 40 of them with CRC, 12 with adenomatous polyps and 32 controls (Table 1). The mean age of patients with CRC was 64 years (±15.18), with polyps was 66 years (±12.37) and for controls 58 years (±12.30).
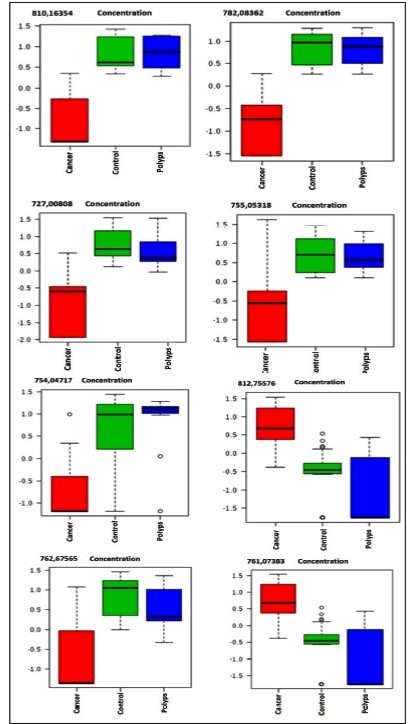
The analysis of boxplot evidenced the distribution of eight VIP ions in the three groups (Table 2). All these ions were in different concentrations in CRC compared to controls or polyps. However, there was no difference between polyps and controls. (Figure 1). Due to the lack of difference between polyps and controls, all analyzes were performed between the cancer and control groups.
Boxblot of the eight ions that differentiated the groups studied, cancer-pre (red), control (green), polyp (blue).
The cluster confirms the difference between the groups (Figure 2).
Lipid spectra heat map comparing the cancer group to control/polyp. The heat map confirms the difference between the cancer and control group.
Initially, 15 ions (m/z) highlighted by PLS-DA were obtained as important lipids to differentiate CRC patients from the others. The concentration between cancers patients were at least 2-fold higher. Most ions were hypo-represented in cancer patients compared to controls and polyps (Figure 3A and 3B).
A) 15 VIPs identified by PLS-DA. The colored boxes on the right indicate the relation of the concentrations of the corresponding metabolite in each of the groups, cancer and control. PLSD-DA Shows Groups Separation According Components 1(96.9%). B) The PCA analysis indicates intrinsic differences between the groups, which could be observed by MALDI-TOF-MS metabolomics profiling.
The ratio between the lipids that best differentiate the groups was 71,587,337 / 76,308,826 with sensitivity of 100% and specificity of 78.6%. The concentration of lipids of patients with tumors that had positive lymph node invasion (N1 or N2) was different from the patients with negative lymph node invasion (Figure 4).
The ROC and Box-Plot curve analysis shows AUCs, a variable that indicates the accuracy of the absence of lymph node invasion (N0) compared to the presence of lymph node invasion (N1 or N2).
DISCUSSION
Lipids play many essential roles in cellular functions, such as survival, proliferation and death. They are involved in the storage of chemical energy, cell signaling, cell membranes and cell-cell interactions in tissues. These cellular processes are strongly related to pathways of carcinogenesis, particularly transformation, progression and metastasis1414. Perrotti F, Rosa C, Cicalini I, Sacchetta P, Del Boccio P, Genovesi D, Pieragostino D. Advances in Lipidomics for Cancer Biomarkers Discovery. Int J Mol Sci . 2016;17(12)..
In our study, we investigated the plasma lipid profile of patients with CRC without metastases and compared to controls and patients with polyps. The analysis tool was composed by an analytical method of MALDI-TOF mass spectrometry to identify possible lipid biomarkers for diagnosis.
Statistical data processing proceeded using the analysis of the main components PCA and the values of the variable importance in the projection (VIP) of the PLS-DA. The PCA is an unsupervised method; each of the main components detects the directions of greater variance in the data matrix, while the PLS-DA is a predictive supervised method of statistical validation for biomarkers1515. Bijlsma S, Bobeldijk I, Verheij ER, Ramaker R, Kochhar S, Macdonald IA, et al. Large-scale human metabolomics studies: a strategy for data (pre-) processing and validation . Anal Chem . 2006;78:567-74..
We observed a different mass spectrum in CRC patients. The small size of the polyps, the low degree of atypia and the reduced number of patients may be the cause of the lack of lipid difference between this group and controls.
We identified eight lipids that best represent the cancer group compared to the control group. The analysis of the ROC curve made with these lipids showed a high accuracy, sensitivity and specificity, suggesting that they may be potential biomarkers. As a way of improving these results, we propose a multiple logistic regression model composed with the eight selected metabolites and obtained the following predictive model.
Our results identified that the ions 810,16354, 782,08362; 754,04717; 727.00808; 755,05318; 761.07383; 762.67565 were hypo-represented and the ion 812,75576 m/z, hyper-represented in the cancer group, indicating the possible association of the lipids with the carcinogenic and metabolic pathways involved in carcinogenensis.
When assessing lymph node involvement, the boxplot test showed that patients with stage III tumors had difference on lipid profiles compared to the others (AUC: 0.924, CI 95%: 0.815-0.982), indicating a high sensitivity and specificity. These results suggest that the ions 71,587,337 and 76,308,826 may be potential prognostic biomarkers.
The increase of lipogenic expression in cancer cells is a common event for the development of cancer, due to rapid cell proliferation and reprogramming of energy metabolism, causing a decrease in plasma lipid levels. According to Corominas-Faja et al., the lipogenic phenotype plays a fundamental role in cellular processes, associated with transformations of cancer cells, including signal transduction, gene expression and response to therapy1616. Corominas-Faja B, Cuyàs E, Gumuzio J, Bosch-Barrera J, Leis O, Martin ÁG, et al. Chemical inhibition of acetyl-CoA carboxylase suppresses self-renewal growth of cancer stem cells. Oncotarget. 2014;30:8306-16..
The lipids were identified according to their categories and classes in polyketide (PK), fatty acids (FA) and glycerolphosphophosphates (GP). The ion 812,5576, hyper-represented in the cancer group, belongs to the category of FA, subclass N-acyl-entolamine1717. de Figueiredo Junior AG, Serafim PVP, de Melo AA, Felipe AV, Lo Turco EG, et al. Analysis of the lipid profile in patients with colorectal cancer in advanced stages. Asian Pac J Cancer Prev. 2018;19:1287-93.. These authors investigated the plasma lipid profile in patients with advanced CRC by MALDI-TOF. They also identified that GP and PK, were hypo represented in the cancer group and hyper represented in the control.
Glycerophospholipids, characterized as phospholipids, are integrators of cell membranes and play the role in cell metabolism and signaling. Demonstrating a relationship of these lipids with metabolic dysregulation and differentiation, certain lipid species strongly linked to cell signaling.
The phospholipid molecules are abundant in the plasma and heterogeneous, this fact is attributed, largely of variations of the FA, as well as the structural differences presented by them. Its structural diversity suggests critical involvement in many physiological and pathological processes1818. Wenk MR, Lipidomics: New Tools and Applications. Cell. 2010;143:888-95.. Prospective studies associated high circulating choline concentration with an increased risk of CRC. Phosphatidylcholines and phosphatidylethanolamines are key components of cell membranes responsible for maintaining structural integrity1919. Shu X, Xiang YB, Rothman N, Yu D, Li HL, Yang G, et al. Prospective study of blood metabolites associated with colorectal cancer risk . Int J Cancer. 2018;143:527-34..
Several prospective studies suggest an association between high intake of flavonoids, particularly flavonols, anthocyanins and a decrease risk of CRC through various anti-inflammatory mechanisms, anti-oxidant properties, induction of apoptosis, inhibition of proliferation and angiogenesis2020. Nimptsch K, Zhang X, Cassidy A, Song M, O’Reilly ÉJ, Lin JH, et al. Habitual Intake of flavonoid subclasses and risk of colorectal cancer in 2 large prospective cohorts. Am J Clin Nutr. 2016;103:184-91.. Protective activities have been widely attributed to anthocyanins by negative regulation of inflammatory signaling pathways, including the light chain of NF-kB, mitogen-activated protein kinase (MAPK), c-Jun N -terminal kinase (JNK), and transducer and transcription activator (STAT). They inhibit cell proliferative pathways, such as the Wnt signaling pathway, which is upregulated in most sporadic CRC. However, the mechanisms that can suppress the colon carcinogenesis has not yet been fully elucidated2121. Li Y, Zhang T, Chen GY. Flavonoids and Colorectal Cancer Prevention. Antioxidants (Basel). 2018;7(12)..
FAs have a structural role in cell membranes, influencing their fluidity, functions and cell membrane composition, as well as being the main structural component of each lipid group. Lipid metabolism is associated with carcinogenicity, progression and alterations in the expression of FA. The lipogenesis process is a known feature that may be present in malignant cells. A key enzyme, fatty acid synthase (FASN) that regulates the biosynthesis of lipogenesis de novo in many cancers, including CRC, is associated with disease aggressiveness and a poor prognosis. Suggesting that this enzyme may be a potential biomarker to investigate the relationships between lipid metabolism and cancer2222. Zaytseva YY, Harris JW, Mitov MI, Kim JT, Butterfield DA, Lee EY, et al. Increased expression of fatty acid synthase provides a survival advantage to colorectal cancer cells via upregulation of cellular respiration. Oncotarget. 2015;6:18891-904.,2323. Notarnicola M, Caruso MG, Tutino V, De Nunzio V, Gigante I, De Leonardis G, et al. Nutrition and lipidomic profile in colorectal cancers. Acta Biomed. 2018;89(9-S):87-96..
Fernandes Messias MC et al., 2018 described reduced levels of different forms of lysophospholipideocolin (LPC), a category of glycerolphospholipid (GP) in plasma of CRC patients. Phospholipids (PLs) play important cellular functions, such as signal transduction, post-translational modifications, homeostasis, adhesion, migration, apoptosis and neurotransmission2424. Fernandes Messias MC, Mecatti GC, Figueiredo Angolini CF, Eberlin MN, Credidio L, Real Martinez CA, et al. Plasma Lipidomic Signature of Rectal Adenocarcinoma Reveals Potential Biomarkers. Front Oncol. 2018;7:325.. In the presence of cancer these molecules are often subject to modifications, with marked changes in the metabolism of phosphatidylcholine (PC) and phosphatidylethanolamine (PE) due to changes in the activity of degradative enzymes, which may be useful for the diagnosis and treatment of cancer2525. Zhao Z, Xiao Y, Elson P, Tan H, Plummer SJ, Berk M, et al. Plasma lysophosphatidylcholine levels: Potential biomarkers for colorectal cancer. J Clin Oncol. 2007;25:2696-701..
Yu Z et al., 2018 evaluated the lipid profiles of 199 patients with lung cancer (NSCLC) and 147 controls by MALDI. The authors defined a panel of four lipid markers with sensitivity of 81.9% and specificity of 70%; that can be lipid biomarkers for the diagnosis of lung carcinomas2626. Yu Z, Chen H, Zhu Y, Ai J, Li Y, Gu W, et al. Global lipidomics reveals two plasma lipids as novel biomarkers for the detection of squamous cell lung cancer: a pilot study. Oncol Lett. 2018;16:761-8..
The role of lipid synthesis in cancer metabolism and tumor development as well as in precancerous lesions still requires many studies to elucidate the numerous mechanisms and pathways of its development. IN clinical practice, the lipid profile is measured based on the serum concentrations of total cholesterol, high-density lipoprotein, low-density lipoprotein and triacylglycerols being poor information for the analysis of the lipid fractions. The mass spectrometry (lipid) technique allowed identifying the specific categories of lipids. Cancer cells are characterized by changes in lipid metabolism. Membrane alterations, rupture of energetic homeostasis, cell signaling, gene expression and protein distribution, affecting several cellular functions, such as apoptosis, autophagy, necrosis, proliferation, differentiation, growth, drug resistance2727. Pakiet A, Kobiela J, Stepnowski P, Sledzinski T, Mika A. Changes in lipids composition and metabolism in colorectal cancer: a review. Lipids Health Dis. 2019;18:29..
In conclusion, we found different lipid profiles between the non-metastatic cancer group and the control group. Eight VIP potential lipid biomarkers were identified.
REFERENCES
-
1Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortalty worldwide for 36 cancers in 185 countries CA Cancer J Clin. 2018;68:394-424.
-
2National Cancer Institute (INCA). Estimate 2018: Cancer incidence in Brazil. [Internet]. [Acessed 2019 June 19]. Available from: https://www.inca.gov.br/search/conteudo/estimativa
» https://www.inca.gov.br/search/conteudo/estimativa -
3Dumoulin FL Hildenbrand R. Endoscopic resection techniques for colorectal neoplasia: current developments. World J Gastroenterol. 2019;25:300-7.
-
4Issa IA, Noureddine M. Colorectal cancer screening: an updated review of the available options. World J Gastroenterol . 2017;23:5086-96.
-
5Bray C, Bell LN, Liang H, Collins D, Yale SH. Colorectal Cancer Screening. WMJ. 2017;116:27-33.
-
6Yiu AJ, Yiu CY. Biomarkers in Colorectal. Anticancer Res. 2016;36:1093-102.
-
7Wang C, Miao Wang M, Han X. Applications of Mass Spectrometry for Cellular Lipid Analysis. Mol Biosyst. 2015;11:698-713.
-
8Li M, Yang L, Bai Y, Liu H. Analytical Methods in Lipidomics and Their Applications. Anal Chem. 2014;86:161-75.
-
9Ingólfsson HI, Melo MN, van Eerden FJ, Arnarez C, Lopez CA, Wassenaar TA, et al. Lipid organization of the plasma membrane. J Am Chem Soc. 2014;136:14554-9.
-
10Li L, Han J, Wang Z, Liu J, Wei J, Xiong S, et al. Mass spectrometry methodology in lipid analysis. Int J Mol Sci, 2014;15:1492-507.
-
11Calvano CD, van der Werf ID, Palmisano F, Sabbatini L. Identification of lipid- and protein-based binders in paintings by direct on-plate wet chemistry and matrix-assisted laser desorption ionization mass spectometry. Anal Bioanal Chem. 2015;407:1015-22.
-
12Takats Z, Strittmatter N, McKenzie JS. Ambient Mass Spectrometry in Cancer Research. Adv Cancer Res. 2017;134:231-56.
-
13Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1957;37:911-7.
-
14Perrotti F, Rosa C, Cicalini I, Sacchetta P, Del Boccio P, Genovesi D, Pieragostino D. Advances in Lipidomics for Cancer Biomarkers Discovery. Int J Mol Sci . 2016;17(12).
-
15Bijlsma S, Bobeldijk I, Verheij ER, Ramaker R, Kochhar S, Macdonald IA, et al. Large-scale human metabolomics studies: a strategy for data (pre-) processing and validation . Anal Chem . 2006;78:567-74.
-
16Corominas-Faja B, Cuyàs E, Gumuzio J, Bosch-Barrera J, Leis O, Martin ÁG, et al. Chemical inhibition of acetyl-CoA carboxylase suppresses self-renewal growth of cancer stem cells. Oncotarget. 2014;30:8306-16.
-
17de Figueiredo Junior AG, Serafim PVP, de Melo AA, Felipe AV, Lo Turco EG, et al. Analysis of the lipid profile in patients with colorectal cancer in advanced stages. Asian Pac J Cancer Prev. 2018;19:1287-93.
-
18Wenk MR, Lipidomics: New Tools and Applications. Cell. 2010;143:888-95.
-
19Shu X, Xiang YB, Rothman N, Yu D, Li HL, Yang G, et al. Prospective study of blood metabolites associated with colorectal cancer risk . Int J Cancer. 2018;143:527-34.
-
20Nimptsch K, Zhang X, Cassidy A, Song M, O’Reilly ÉJ, Lin JH, et al. Habitual Intake of flavonoid subclasses and risk of colorectal cancer in 2 large prospective cohorts. Am J Clin Nutr. 2016;103:184-91.
-
21Li Y, Zhang T, Chen GY. Flavonoids and Colorectal Cancer Prevention. Antioxidants (Basel). 2018;7(12).
-
22Zaytseva YY, Harris JW, Mitov MI, Kim JT, Butterfield DA, Lee EY, et al. Increased expression of fatty acid synthase provides a survival advantage to colorectal cancer cells via upregulation of cellular respiration. Oncotarget. 2015;6:18891-904.
-
23Notarnicola M, Caruso MG, Tutino V, De Nunzio V, Gigante I, De Leonardis G, et al. Nutrition and lipidomic profile in colorectal cancers. Acta Biomed. 2018;89(9-S):87-96.
-
24Fernandes Messias MC, Mecatti GC, Figueiredo Angolini CF, Eberlin MN, Credidio L, Real Martinez CA, et al. Plasma Lipidomic Signature of Rectal Adenocarcinoma Reveals Potential Biomarkers. Front Oncol. 2018;7:325.
-
25Zhao Z, Xiao Y, Elson P, Tan H, Plummer SJ, Berk M, et al. Plasma lysophosphatidylcholine levels: Potential biomarkers for colorectal cancer. J Clin Oncol. 2007;25:2696-701.
-
26Yu Z, Chen H, Zhu Y, Ai J, Li Y, Gu W, et al. Global lipidomics reveals two plasma lipids as novel biomarkers for the detection of squamous cell lung cancer: a pilot study. Oncol Lett. 2018;16:761-8.
-
27Pakiet A, Kobiela J, Stepnowski P, Sledzinski T, Mika A. Changes in lipids composition and metabolism in colorectal cancer: a review. Lipids Health Dis. 2019;18:29.
-
Disclosure of funding: The study had been supported by CAPES Coordination for the Improvement of Higher Education Personnel.
Publication Dates
-
Publication in this collection
28 Nov 2019 -
Date of issue
Oct-Dec 2019
History
-
Received
29 July 2019 -
Accepted
27 Sept 2019