ABSTRACT
BACKGROUND:
Colorectal cancer is the third most common neoplasm in the world. Methylation of tumor related genes in CpG islands can cause gene silencing and been involved in the development of cancer. The potential role of DKK2 as a biomarker for early diagnosis of colorectal cancer remains unclear.
OBJECTIVE:
The aim of the study was to evaluate the profile of methylation and RNAm expression of DKK2 as potential predictors of colorectal cancer diagnosis and prognosis.
METHODS:
Expression of mRNAs encoding DKK2 in 35 colorectal cancer tissues was quantified using real-time polymerase chain reaction analysis. The DNA methylation was studied by high resolution melting analysis. The general characteristics of the patients were collected. DKK2 methylation and expression were compared to clinical, pathological aspects and overall survival.
RESULTS:
Among the 35 patients studied, 18 were male, 10 were on right colon and 25 on left colon. Among the 20 patients with high hypermethylation, 15 of them had mRNA low expression of DKK2. There was no significant association between DKK2 promoter methylation and mRNA DKK2 expression and clinical or pathological features. DKK2 promoter methylation (P=0.154) and DKK2 RNA expression (P=0.345) did not show significant correlation with overall survival.
CONCLUSION:
DKK2 promoter methylation and DKK2 RNA status appear to be biomarkers of cancer diagnosis but not predictors of prognosis.
HEADINGS:
Colorectal neoplasms; Tumor biomarkers; Methylation; Gene silencing; Histocytochemistry
RESUMO
CONTEXTO:
O câncer colorretal é a terceira neoplasia mais comum no mundo. A metilação de alguns genes nas ilhas CpG podem causar silenciamento gênico e estar envolvida no desenvolvimento de câncer. O potencial papel de DKK2 como um biomarcador no diagnóstico precoce de CCR permanece incerto.
OBJETIVO:
O objetivo do estudo foi avaliar o perfil de metilação e expressão de RNAm do gene DKK2 para identificar preditores potenciais de diagnóstico e prognóstico de CCR.
MÉTODOS:
A expressão de mRNAs que codificam DKK2 em 35 tecidos de câncer colorretal foi quantificada por reação em cadeia da polimerase em tempo real e a metilação do DNA foi verificada por análise de alta resolução. As características gerais dos pacientes foram coletadas. A metilação e expressão de DKK2 foram comparadas aos aspectos clínicos, patológicos e à sobrevida global.
RESULTADOS:
Entre os 35 pacientes estudados, 18 eram do sexo masculino, 10 tumores eram do cólon ascendente ou transverso e 25 do descendente ou reto. Entre os 20 pacientes com hipermetilação, 12 deles apresentaram baixa expressão de RNAm do gene DKK2. Não houve associação significativa entre a metilação do promotor de DKK2 e a expressão de RNAm de DKK2 e características clínicas ou patológicas. A metilação do promotor de DKK2 e a expressão do RNA de DKK2 não mostraram correlação com sobrevida global dos pacientes com CCR.
CONCLUSÃO:
A metilação do gene promotor e a expressão do RNAm do gene DKK2 parecem ser biomarcadores de diagnóstico de câncer, mas não se mostraram úteis na avaliação prognóstica.
DESCRITORES:
Neoplasias colorretais; Biomarcadores tumorais; Metilação; Inativação gênica; Histocitoquímica
INTRODUCTION
Colorectal cancer (CRC) is the most common malignant neoplasm of the gastrointestinal tract, with approximately 1,800,000 new cases diagnosed yearly11. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: Cancer J Clin. 2018;68:394-424. (Bray et al.), being the third cause of cancer. In Brazil, CRC is the second cause of cancer in men and women22. INCA National Institute of Cancer: Estimate/2020 - Cancer Incidence in Brazil. Available from: https://rbc.inca.gov.br/revista/index.php/revista/article/view/927
https://rbc.inca.gov.br/revista/index.ph...
(INCA, 2020). According to Globocan, in 2018, there were more than 1.8 million new cases of colorectal cancer (CRC) with 881 thousand deaths.
Ageing with more than 50 years, high ingestion of fat and low in fiber, tobacco, inflammatory bowel disease and presence of adenomas are considered risk factors for CRC33. Aleksandrova K, Pischon T, Jenab M, Bueno-de-Mesquita HB, Fedirko V, Norat T, et al., Combined impact of healthy lifestyle factors on colorectal cancer: a large European cohort study. BMC Med. 2014;2:168..
DNA alterations may be inherited in 15-20%44. Hemminki K, Chen B. Familial risk for colorectal cancers are mainly due to heritable causes. Cancer Epidemiol Biomarkers Prev. 2004;13:1253-6. of the patients or caused mainly by interaction of environment and lifestyle33. Aleksandrova K, Pischon T, Jenab M, Bueno-de-Mesquita HB, Fedirko V, Norat T, et al., Combined impact of healthy lifestyle factors on colorectal cancer: a large European cohort study. BMC Med. 2014;2:168.. Several mutations in the same clone of cells transform normal epithelial cells in cancer cells, with increased proliferation and loss of apoptosis and growth control.
Early screening denotes a central importance in the diagnosis of adenomas and early CRC55. Winawer SJ. Colorectal cancer screening. Best Pract Res Clin Gastroenterol. 2007;21:1031-48.. There are a large number of studies focused on finding potential biomarkers through promoter methylation of tumor-related genes and expression of tumor suppressor genes in gastrointestinal cancers and precancerous lesions, suggesting their involvement in the progression of multiple levels of colorectal carcinogenesis66. Wang C, Yue Y, Shao B, Qiu Z, Mu J, Tang J, et al. Dickkopf -related Protein 2 is epigenetically inactivated and suppresses colorectal cancer growth and tumor metastases by antagonizing Wnt/β-Catenin signaling. Cell Physiol Biochem. 2017;41:1709-24..
High methylation of tumor related genes in CpG islands can cause gene silencing and been involved in the development of cancer66. Wang C, Yue Y, Shao B, Qiu Z, Mu J, Tang J, et al. Dickkopf -related Protein 2 is epigenetically inactivated and suppresses colorectal cancer growth and tumor metastases by antagonizing Wnt/β-Catenin signaling. Cell Physiol Biochem. 2017;41:1709-24.,77. Okugawa Y, Grady WM, Goel A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology. 2015;149:1204-25.. Alterations of the Wnt/β-catenin signaling pathway are common in tumorigenesis of CRC and epigenetic modifications may be involved in the regulation of this pathway cancer88. Kazanets A, Shorstova T, Hilmi K, Marques M, Witcher M. Epigenetic silencing of tumor suppressor genes: Paradigms, puzzles, and potential. Biochim Biophys Acta. 2016;1865:275-88.,99. Lee SK, Hwang JH, Choi KY. Interaction of the Wnt/β-catenin and RAS-ERK pathways involving co-stabilization of both β-catenin and RAS plays important roles in the colorectal tumorigenesis. Adv Biol Regul. 2018;68:46-54..
The Dickkopf family (DKK) is composed by four glycoproteins (DKK1-4), secreted with two cysteine-rich domains, separated by a binding region. DKK2 is a Wnt signaling inhibitor that is generally supraregulated in human cancers, including CRC. Studies on expression and methylation of DKK2 in CRC suggest that DKK2 functions as a tumor suppressor and Wnt / β-catenin pathway regulator66. Wang C, Yue Y, Shao B, Qiu Z, Mu J, Tang J, et al. Dickkopf -related Protein 2 is epigenetically inactivated and suppresses colorectal cancer growth and tumor metastases by antagonizing Wnt/β-Catenin signaling. Cell Physiol Biochem. 2017;41:1709-24..
A study already published in CRC fresh tumor tissue, by our group showed that all tumors cancer was methylated compared to normal tissues1010. Silva TD, Vidigal VM, Felipe AV, De Lima JM, Neto RA, Saad SS, et al. DNA methylation as an epigenetic biomarker in colorectal cancer. Oncol Lett. 2013;6:1687-92. (Silva). However, the circulating free DNA (cfDNA) of DKK2 in peripheral blood of these patients was increased in only 25% of them1111. Silva TD, Felipe AV, Pimenta CA, Barão K, Forones NM. DNA methylation profile of the DKK2 gene as a biomarker in patients with colorectal cancer. Genet Mol Res. 2012;11:3138-45. doi: 10.4238/gmr16039816
https://doi.org/10.4238/gmr16039816...
.
The main goal of this study was to evaluate DKK2 gene expression and methylation as a biomarker of diagnosis and prognosis in colorectal tumors and analyze the influence of DKK2 methylation in mRNA expression.
METHODS
The study was approved by the Ethical Committee of the Federal University of São Paulo (CAAE no 58914716.0.0000.5505) and all subjects have given their informed written consent. The 35 biological samples were obtained during surgical resection of the tumor at Hospital Sao Paulo, UNIFESP, Brazil. Patients with familiar history of cancer, inflammatory bowel disease, radiotherapy, chemotherapy treatment, or concurrent or history of other types of cancers were excluded. DNA and RNA was isolated from the surgically removed colorectal cancer tumors.
Extraction of genomic DNA
Around 25 mg of each tumor tissue was mechanically disrupted using a Tissue Ruptor® (Qiagen, Venlo, Netherlands). The genomic DNA extracted and purified from fresh-frozen colorectal cancer samples using QIAmp DNA Mini kit (Qiagen, Milan, Italy) according to the manufacturer’s instruction. The extracted DNA was measured using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
Bisulfite modification
Bisulfite conversion of isolated 200ng of genomic DNA from each sample were treated with sodium bisulfite using Cells-to-CpG™ Bisulfite Conversion Kit (Thermo Scientific, Wilmington, USA), according to the manufacturer’s protocol. Briefly, Sodium bisulphite converts unmethylated cytosines to uracils, whilst 5-methylcytosines are left unchanged.
DNA methylation by high resolution melting (HRM) analysis
For the MS-HRM of DKK2 gene we used methylation independent primers (MIP), based on previous study1111. Silva TD, Felipe AV, Pimenta CA, Barão K, Forones NM. DNA methylation profile of the DKK2 gene as a biomarker in patients with colorectal cancer. Genet Mol Res. 2012;11:3138-45. doi: 10.4238/gmr16039816
https://doi.org/10.4238/gmr16039816...
. Briefly, primers for the DKK2 gene were designed to amplify methylated DNA and unmethylated DNA using Methyl Primer Express v 1.0 (Applied Biosystems, Foster City, CA, USA). The primer sequences for DKK2 HM forward and reverse was AGGTATCGTTGCGTTGGTAGC and AAATCAAAAAACGTCCCCGA respectively. The primer sequences for DKK2 HM forward and reverse was UM forward and reverse respectively was TTTAGGTATTGTTGTGTTGGTAGT and AAATCAAAAAACATCCCCAAACC.
The PCR analyses were run using the following conditions: one cycle of 95°C for 15 min, 60 cycles of 95°C for 15 s, 63°C annealing temperature for 30 s and 72°C for 30 s; followed by an HRM step of 95°C for 10 s and 50°C for 1 min, 70°C for 15 s, and continuous acquisition to 95°C at one acquisition per 0.1°C. The PCR reaction was set up in a final volume of 20 µL, containing 10-12,5 µL of MeltDoctor HRM master mix (Applied Biosystems, Foster City, CA USA), 10 pmol of each primer and 1 µL (almost 10-20 ng) of bisulfite modified DNA template. The experiments were performed in triplicate. Figure 1 shows the melting profiles of the two promoter regions analyzed. Control DNAs with varying the ratios of methylation: 0%, 25%, 50%, 75% and 100%, were prepared by mixing following the manufacturer’s protocol (EpiTect methylated and unmethylated human control DNA, bisulfite converted, Qiagen, Milan, Italy). A standard curve with known methylation ratios was included in each assay to provide a quantitative analysis of DNA methylation profile of the promoter of DKK2 gene. The profile of the aligned fluorescence intensity and melting temperature for serial dilutions of methylated DNA for the DKK2 gene were plotted in the melting graph (Figure 1). The tumors with more than 25% of methylation were considered hypermethylated.
Fluorescence intensity profile aligned to the melting temperature for serial dilution of methylated DNA for the DKK2 gene.
The reported relative fluorescence units (RFU) value within the melt curve was exported to Microsoft Excel and RFU average of each DNA standard was then used to obtain an interpolation curve. Finally, imputation of the RFU value (corresponding to the analyzed sample) to the polynomial function provided a precise (single value) estimate of the percentage of methylation.
Quantitative real-time reverse transcription PCR (qRT-PCR)
Reverse transcription reaction was performed using the Superscript VILO cDNA synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) following the manufacturer’s protocol. Expression assays were performed by using TaqMan Gene Expression Assays for human DKK2 under the inventory ID: Hs00205294_m1 [Amplicon Length: 88pb]. Human Endogenous control assays ACTB (encoding for beta actin, assay ID: Hs01060665_g1, amplicon length: 63pb) and GAPDH (encoding for glyceraldehyde-3-phosphate dehydrogenase, ID: Hs02786624_g1, amplicon length: 157pb) (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) served as references for normalization. TaqMan Gene Expression Master Mix according to the manufacturer’s protocol (Applied Biosystems, Carlsbad, CA, USA). Reverse transcriptase reaction and Real-Time PCR were performed using the Step one plus Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). Briefly, the reactions were incubated in a 96-well plate at 95°C for 20 s, followed by 40 cycles of 95°C for 1 s and 60°C for 20 s. In each experiment, triplicate reactions were performed in each sample. Lastly, the relative expression was calculated as the ratio of DKK2 to ACTB for each sample (calculated as the mean of triplicate). The relative quantity of mRNA for each sample were calculated by 2−ΔΔCT, where ΔCT = (mean of triplicate CTABCB1 − mean of triplicate CTACTB ) and ΔΔCT = (ΔCT - mean ΔCT of all the samples). In pilot studies, the PCR amplicons were evaluated on agarose gels to verify product size.
Statistical analysis
The polynomial function determined by the polynomial regression curve of standards method estimates the percentage of methylation in each experiment. The measurements were performed in triplicate and data are expressed as mean ± standard deviation (SD). Pearson’s correlation analysis was used to determine the relationship between mRNA DKK2 expressions and DNA methylation profile of the promoter of DKK2 gene. All other statistical differences were determinate with unpaired Student’s t-test or one-way ANOVA followed by the Tukey test for multiple comparisons. Median survival was based on Kaplan-Meier curves and tested for significance by the log-rank test. The statistical analysis was performed using the Minitab V16.0 (State College, PA), or SPSS V 23 (SPSS Inc., Chicago, IL) or Office Excel for Windows 2010 (Microsoft, Redmond, WA). P value of <0.05 was considered statistically significant.
RESULTS
Thirty-five patients with colorectal tumors with 63.6±13.9 years, range 47-78 years, being 18 male and 20 with stage III or IV tumors were studied. Among these 10 were of right colon and 25 of left colon or rectum, called left colon.
This investigation extended the DNA methylation analysis of our previous study1111. Silva TD, Felipe AV, Pimenta CA, Barão K, Forones NM. DNA methylation profile of the DKK2 gene as a biomarker in patients with colorectal cancer. Genet Mol Res. 2012;11:3138-45. doi: 10.4238/gmr16039816
https://doi.org/10.4238/gmr16039816...
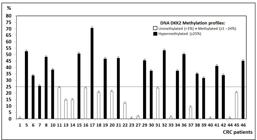
, that studied DNA methylation of DKK2 gene by HRM method. The DKK2 gene promoter unmethylated/hypomethylated and hypermethylated status are shown in Figure 2. Among the tumors 35 tumors, 20 are hypermethylated.
The relative quantity of mRNA expression was normalized to the relative quantity of ACTB for each sample and performed by qRT-PCR. The DKK2 gene expression status is shown on Figure 3. Among the 35 tumors, 24 had low expression of RNAm of DKK2. Low expression of this gene and hypermethylation was observed in 15 (43%) patients.
The DKK2 DNA methylation status and relative quantity of mRNA expressions and their relationship with clinical and pathological features in CRC patients are showed in Table 1. There is no significant association between genes methylation status and age, sex, tumor site, TNM stage and metastasis. There is also no significant association between DKK2 expression and clinical or pathological features (Table 2).
The results show that the expression level of the DKK2 mRNA was not correlated with the increasing or decreasing percentage of methylated DKK2 gene in methylated samples.
The patients were followed for a period of 4.9±2.5 years. There was no difference on survival for DKK2 promoter methylation (P=0.154) or DKK2 mRNA expression status (P=0.345).
DISCUSSION
The number of world CRC will increase during the next years, although in some countries the screening methods caused a decrease incidence in individuals with more than 50 years or earlier in patients with family history of CRC. In contrast, the number of younger patients with CRC are increasing with not elucidation of the cause1212. Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, et al. Colorectal cancer statistics, 2017. CACancer J Clin . 2017;67:177-193..
Between the 35 patients with CRC included, 51.4% were men, the average age was 63.3±13.9 years, and most patients were over the age of 50 years (82.9%). These data are consistent with those published by INCA, which describes the incidence of CRC in Brazil2 (INCA, 2020). The limitations of the study were the short number of tissues evaluated and the absence of adjacent free margin.
The DKK2 gene is deleted in some cancers, but epigenetic alterations causing the methylation of the promoter region of the CpG islands may cause gene silencing that occurs by DNA binding proteins, a blocking of transcriptional regulators and a consequent decreased transcription1313. Lopez-Serra L, Esteller M. Proteins that bind methylated DNA and human cancer: Reading the wrong words. Br J Cancer. 2008;98:1881-5..
Although the molecular mechanism of DKK2 is not well defined, some authors suggest that DKK2 functions as a tumor suppressor gene and a regulator of the Wnt/β-catenin pathway in CRC66. Wang C, Yue Y, Shao B, Qiu Z, Mu J, Tang J, et al. Dickkopf -related Protein 2 is epigenetically inactivated and suppresses colorectal cancer growth and tumor metastases by antagonizing Wnt/β-Catenin signaling. Cell Physiol Biochem. 2017;41:1709-24.,77. Okugawa Y, Grady WM, Goel A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology. 2015;149:1204-25.. Maehata et al., demonstrated in gastrointestinal cancer cell lines that CpG island hypermethylation is associated with gene silencing1414. Maehata T, Taniguchi H, Yamamoto H, Nosho K, Adachi Y, Miyamoto N, et al. Transcriptional silencing of Dickkopf gene family by CpG island hipermethylation in human gastrointestinal cancer. World J Gastroenterol. 2008;14:2702-14.,1515. Bae JM, Kim JH, Kang GH. Epigenetic alterations in colorectal cancer: the CpG island methylator phenotype. Histol Histopathol. 2013;28:585-95..
Hu et al.1616. Hu J, Wang Z, Chen Z, Li Ao, Sun J, Zheng M, et al. DKK2 blockage-mediated immunotherapy enhances anti-angiogenic therapy of Kras mutated colorectal cancer. Biomed Pharmacother 2020;127:110299. described that the blockade of DKK2 by an antibody decreased tumor progression by immune cell activation and tumor angiogenesis suggesting that the combination of anti-DKK2 and anti-VEGFr can be a possible treatment of advancer CRC.
The methylation status of this gene and others is frequently analyzed by PCR, which evaluate qualitatively the presence or absence of methylation by bisulfite conversion-based methods that involve the conversion of non-methylated cytosines to uracil after treatment. This method did not allow the quantification of DNA methylated and cannot classify tissues with low methylation to others with higher methylation. In this assay, the methylation state was studied by HRM, which allows the quantitative determination. Some authors fail to associate methylation status with cancer, and others demonstrate that even methylated genes can continue to perform their functions normally. Based on this information it is important to discuss whether it is sufficient to describe the presence of methylation or whether it is necessary to go further and quantify the percentage1515. Bae JM, Kim JH, Kang GH. Epigenetic alterations in colorectal cancer: the CpG island methylator phenotype. Histol Histopathol. 2013;28:585-95.,1717. Hrašovec S, Hauptman N, Glavač D, Jelenc F, Ravnik-Glavac M. TMEM25 is a candidate biomarker methylated and down-regulated in colorectal cancer. Dis Markers. 2013;34:93-104.,1818. Nilsson TK, Löf-Öhlin ZM, Sun XF. DNA methylation of the p14ARF, RASSF1A and APC1A genes as an independent prognostic factor in colorectal cancer patients. Int J Oncol. 2013;42:127-33..
DNA hypermethylation of DKK2 was observed in 19 (54.3%) tumors and hipomethylated or unmethylated in the others (Table 1). In another study described by our group the hypermethylation of DKK2 was described in 63% of the tumors tissues, but only in 4.45% of the patients with normal colonoscopy (P<0.001)1111. Silva TD, Felipe AV, Pimenta CA, Barão K, Forones NM. DNA methylation profile of the DKK2 gene as a biomarker in patients with colorectal cancer. Genet Mol Res. 2012;11:3138-45. doi: 10.4238/gmr16039816
https://doi.org/10.4238/gmr16039816...
. The highest proportion of DKK2 hypermethylated in CRC found in this study and in the Silva et al.1111. Silva TD, Felipe AV, Pimenta CA, Barão K, Forones NM. DNA methylation profile of the DKK2 gene as a biomarker in patients with colorectal cancer. Genet Mol Res. 2012;11:3138-45. doi: 10.4238/gmr16039816
https://doi.org/10.4238/gmr16039816...
, suggest that this gene is a biomarker of cancer. Jones and Takai found that patients with methylation of DNA in normal areas adjacent to the tumor had worst prognosis1919. Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068-70..
Epigenetic alterations occur during aging process and may be an explanation for the higher incidence of CRC after 60 years old. These alterations in older patients were described in human prostate cancer2020. Damaschke NA, Yang B, Bhusari S, Svaren JP, Jarrard DF. Epigenetic Susceptibility Factors for Prostate Cancer with Aging. Prostate. 2013; 73: 1721-1730. and CRC.
The hypermethylation has been described in precancerous adenoma and can be serve as early detection markers2121. Galamb O, Kalmár A, Péterfia B, Csabai I, Bodor A, Ribli D, et al. Aberrant DNA methylation of WNT pathway genes in the development and progression of CIMP-negative colorectal cancer. Epigenetics. 2016;11:588-602.
22. Beggs AD, Jones A, El-Bahrawy M, Abulafi M, Hodgson SV, Tomlinson IP. Whole-genome methylation analysis of benign and malignant colorectal tumours. J Pathol. 2013;229:697-704.-2323. Luo Y, Wong CJ, Kaz AM, Dzieciatkowski S, Carter KT, Morris SM, et al. Differences in DNA methylation signatures reveal multiple pathways of progression from adenoma to colorectal cancer. Gastroenterology. 2014;147:418-29.. In this study, we did not found difference in methylation status and pathological stages. These results reinforced that DNA methylation can serve as early detection markers.
Most of the tumors studied (68.6%) had lower expression of RNAm of DKK2 gene and was not influenced by age, sex, site of the tumor or pathological stage.
Some studies published that hypermethylation of some genes in the promoter region may silence mRNA expression. Molnar et al. observed in several TP53 pathway genes, promoter methylation and RNAm expression alterations2424. Molnár B, Galamb O, Péterfia B, Wichmann B, Csabai I, Bodor A, et al. Gene promoter and exon DNA methylation changes in colon cancer development - mRNA expression and tumor mutation alterations. BMC Cancer 2018;18:695. doi 10.1186/s12885-018-4609-x.
https://doi.org/10.1186/s12885-018-4609-...
.
Although we did not find a correlation between hypermethylation of DNA and RNAm expression, methylation of the promoter gene may be the cause of the lower expression in 43% of the tumors. In these tumors downregulation or silencing of DKK2 expression are associated with the hypermethylation status of its promoter. This fact can suggest the treatment of some cancer patients with demethylation drugs.
There are several molecular differences including DNA methylation between left and right colon2525. Horii J, Hiraoka S, Kato J, Harada K, Kuwaki K, Fujita H, et al. Age-related methylation in normal colon mucosa differs between the proximal and distal colon in patients who underwent colonoscopy. Clin Biochem. 2008;41:1440-8., mainly in tumors with microsatellite instability (MSI) or microsatellite stability (MSS). In the present study, we examined DNA methylation and DKK2 mRNA expression in patients without familiar history of cancer. In these patients, MSI is rare. Hypermethylation (78.9%) and lower expression of DKK2 (79.2%) was most prevalent in distal tumors. A previous study reported that specific cancer-related genes demonstrate differential methylation depending on colon location2626. Sugai T, Habano W, Jiao Y-F, Tsukahara M, Takeda Y, Otsuka K, et al. Analysis of molecular alterations in left- and right-sided colorectal carcinomas reveals distinct pathways of carcinogenesis: proposal for new molecular profile of colorectal carcinomas. J Mol Diagn. 2006;8:193-201. in tumors with MSS. In a recent study2727. Sugai T, Yoshida M, Eizuka M, Uesugii N, Habano W, Otsuka K, et al. Analysis of the DNA methylation level of cancer-related genes in colorectal cancer and the surrounding normal mucosa. Clin Epigenetics. 2017;9:55. doi 10.1186/s13148-017-0352-4.
https://doi.org/10.1186/s13148-017-0352-...
the authors described an increased DNA methylation levels of various genes, including DKK2 in CRC tumors and adjacent mucosa with an MSS phenotype.
Univariate analysis by the Kaplan-Meier curves indicates that DKK2 promoter methylation shows no significant difference between methylation and prognostic factors in CRC patients. Similarly, there is no significant association between DKK2 RNA expression status and overall survival in CRC patients.
In conclusion, the frequency of DKK2 gene hypermethylation and low RNAm expression is frequent on colorectal cancer tissue independent of the stage suggesting that this gene may be a biomarker of cancer. However, hypermethylation or low expression of DKK2 are not markers of prognosis in the tissues examined.
ACKNOWLEDGEMENTS
This research was supported by (Capes) Coordenação de Aperfeiçoamento de Pessoal de Nível Superior and (Fapesp) Fundação de Amparo à Pesquisa do Estado de São Paulo, grant n. 2013/19268-3.
REFERENCES
-
1Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: Cancer J Clin. 2018;68:394-424.
-
2INCA National Institute of Cancer: Estimate/2020 - Cancer Incidence in Brazil. Available from: https://rbc.inca.gov.br/revista/index.php/revista/article/view/927
» https://rbc.inca.gov.br/revista/index.php/revista/article/view/927 -
3Aleksandrova K, Pischon T, Jenab M, Bueno-de-Mesquita HB, Fedirko V, Norat T, et al., Combined impact of healthy lifestyle factors on colorectal cancer: a large European cohort study. BMC Med. 2014;2:168.
-
4Hemminki K, Chen B. Familial risk for colorectal cancers are mainly due to heritable causes. Cancer Epidemiol Biomarkers Prev. 2004;13:1253-6.
-
5Winawer SJ. Colorectal cancer screening. Best Pract Res Clin Gastroenterol. 2007;21:1031-48.
-
6Wang C, Yue Y, Shao B, Qiu Z, Mu J, Tang J, et al. Dickkopf -related Protein 2 is epigenetically inactivated and suppresses colorectal cancer growth and tumor metastases by antagonizing Wnt/β-Catenin signaling. Cell Physiol Biochem. 2017;41:1709-24.
-
7Okugawa Y, Grady WM, Goel A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology. 2015;149:1204-25.
-
8Kazanets A, Shorstova T, Hilmi K, Marques M, Witcher M. Epigenetic silencing of tumor suppressor genes: Paradigms, puzzles, and potential. Biochim Biophys Acta. 2016;1865:275-88.
-
9Lee SK, Hwang JH, Choi KY. Interaction of the Wnt/β-catenin and RAS-ERK pathways involving co-stabilization of both β-catenin and RAS plays important roles in the colorectal tumorigenesis. Adv Biol Regul. 2018;68:46-54.
-
10Silva TD, Vidigal VM, Felipe AV, De Lima JM, Neto RA, Saad SS, et al. DNA methylation as an epigenetic biomarker in colorectal cancer. Oncol Lett. 2013;6:1687-92.
-
11Silva TD, Felipe AV, Pimenta CA, Barão K, Forones NM. DNA methylation profile of the DKK2 gene as a biomarker in patients with colorectal cancer. Genet Mol Res. 2012;11:3138-45. doi: 10.4238/gmr16039816
» https://doi.org/10.4238/gmr16039816 -
12Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, et al. Colorectal cancer statistics, 2017. CACancer J Clin . 2017;67:177-193.
-
13Lopez-Serra L, Esteller M. Proteins that bind methylated DNA and human cancer: Reading the wrong words. Br J Cancer. 2008;98:1881-5.
-
14Maehata T, Taniguchi H, Yamamoto H, Nosho K, Adachi Y, Miyamoto N, et al. Transcriptional silencing of Dickkopf gene family by CpG island hipermethylation in human gastrointestinal cancer. World J Gastroenterol. 2008;14:2702-14.
-
15Bae JM, Kim JH, Kang GH. Epigenetic alterations in colorectal cancer: the CpG island methylator phenotype. Histol Histopathol. 2013;28:585-95.
-
16Hu J, Wang Z, Chen Z, Li Ao, Sun J, Zheng M, et al. DKK2 blockage-mediated immunotherapy enhances anti-angiogenic therapy of Kras mutated colorectal cancer. Biomed Pharmacother 2020;127:110299.
-
17Hrašovec S, Hauptman N, Glavač D, Jelenc F, Ravnik-Glavac M. TMEM25 is a candidate biomarker methylated and down-regulated in colorectal cancer. Dis Markers. 2013;34:93-104.
-
18Nilsson TK, Löf-Öhlin ZM, Sun XF. DNA methylation of the p14ARF, RASSF1A and APC1A genes as an independent prognostic factor in colorectal cancer patients. Int J Oncol. 2013;42:127-33.
-
19Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068-70.
-
20Damaschke NA, Yang B, Bhusari S, Svaren JP, Jarrard DF. Epigenetic Susceptibility Factors for Prostate Cancer with Aging. Prostate. 2013; 73: 1721-1730.
-
21Galamb O, Kalmár A, Péterfia B, Csabai I, Bodor A, Ribli D, et al. Aberrant DNA methylation of WNT pathway genes in the development and progression of CIMP-negative colorectal cancer. Epigenetics. 2016;11:588-602.
-
22Beggs AD, Jones A, El-Bahrawy M, Abulafi M, Hodgson SV, Tomlinson IP. Whole-genome methylation analysis of benign and malignant colorectal tumours. J Pathol. 2013;229:697-704.
-
23Luo Y, Wong CJ, Kaz AM, Dzieciatkowski S, Carter KT, Morris SM, et al. Differences in DNA methylation signatures reveal multiple pathways of progression from adenoma to colorectal cancer. Gastroenterology. 2014;147:418-29.
-
24Molnár B, Galamb O, Péterfia B, Wichmann B, Csabai I, Bodor A, et al. Gene promoter and exon DNA methylation changes in colon cancer development - mRNA expression and tumor mutation alterations. BMC Cancer 2018;18:695. doi 10.1186/s12885-018-4609-x.
» https://doi.org/10.1186/s12885-018-4609-x -
25Horii J, Hiraoka S, Kato J, Harada K, Kuwaki K, Fujita H, et al. Age-related methylation in normal colon mucosa differs between the proximal and distal colon in patients who underwent colonoscopy. Clin Biochem. 2008;41:1440-8.
-
26Sugai T, Habano W, Jiao Y-F, Tsukahara M, Takeda Y, Otsuka K, et al. Analysis of molecular alterations in left- and right-sided colorectal carcinomas reveals distinct pathways of carcinogenesis: proposal for new molecular profile of colorectal carcinomas. J Mol Diagn. 2006;8:193-201.
-
27Sugai T, Yoshida M, Eizuka M, Uesugii N, Habano W, Otsuka K, et al. Analysis of the DNA methylation level of cancer-related genes in colorectal cancer and the surrounding normal mucosa. Clin Epigenetics. 2017;9:55. doi 10.1186/s13148-017-0352-4.
» https://doi.org/10.1186/s13148-017-0352-4
-
Disclosure of funding:Fundação de Amparo à Pesquisa do Estado de São Paulo (Fapesp), São Paulo, SP, Brasil.
Publication Dates
-
Publication in this collection
23 Apr 2021 -
Date of issue
Jan-Mar 2021
History
-
Received
27 July 2020 -
Accepted
28 Sept 2020