Abstracts
More than 70 species of mycobacteria have been defined, and some can cause disease in humans, especially in immunocompromised patients. Species identification in most clinical laboratories is based on phenotypic characteristics and biochemical tests and final results are obtained only after two to four weeks. Quick identification methods, by reducing time for diagnosis, could expedite institution of specific treatment, increasing chances of success. PCR restriction-enzyme analysis (PRA) of the hsp65 gene was used as a rapid method for identification of 103 clinical isolates. Band patterns were interpreted by comparison with published tables and patterns available at an Internet site (http://www.hospvd.ch:8005). Concordant results of PRA and biochemical identification were obtained in 76 out of 83 isolates (91.5%). Results from 20 isolates could not be compared due to inconclusive PRA or biochemical identification. The results of this work showed that PRA could improve identification of mycobacteria in a routine setting because it is accurate, fast, and cheaper than conventional phenotypic identification.
Mycobacterium; PCR; Diagnosis; Identification
Mais de 70 espécies de micobactérias já foram definidas e algumas delas podem causar enfermidade em humanos, especialmente em pacientes imunocomprometidos. A identificação de espécie, na maioria dos laboratórios clínicos, se baseia em características fenotípicas e testes bioquímicos e resultados definitivos só são obtidos após duas a quatro semanas. Métodos rápidos de identificação reduzem o tempo necessário para o diagnóstico e podem antecipar a instituição do tratamento específico, aumentando as chances de sucesso. A análise de padrões de restrição do gene hsp65 amplificado por PCR (PRA) foi utilizada como método rápido de identificação em 103 isolamentos clínicos. Os padrões de bandas foram interpretados por comparação com tabelas publicadas e padrões disponíveis em um site de Internet (<a HREF="http://www.hospvd.ch:8005/">http://www.hospvd.ch:8005</a>). Resultados concordantes de PRA e identificação bioquímica foram obtidos em 76 de 83 isolamentos (91,5%). Os resultados de 20 isolamentos não puderam ser comparados porque a identificação fenotípica ou por PRA foi inconclusiva. Os resultados deste trabalho mostram que PRA pode ser útil para identificação de rotina de micobactérias por ser um método acurado, rápido e mais econômico do que a identificação convencional.
hsp65 PCR-RESTRICTION ENZYME ANALYSIS (PRA) FOR IDENTIFICATION OF MYCOBACTERIA IN THE CLINICAL LABORATORY
Carolina Feher da SILVA(1 (1 ) Departamento de Microbiologia, Imunologia e Parasitologia - Universidade Federal de São Paulo, Escola Paulista de Medicina (UNIFESP-EPM). Rua Botucatu 862, 3° andar, 04023-062 São Paulo, SP, Brasil. (2 ) Setor de Micobactérias, Instituto Adolfo Lutz de São Paulo. Av. Dr. Arnaldo 351, 9 o andar, 01246-902 São Paulo, SP, Brasil. ), Suely Yoko Mizuka UEKI(2 (1 ) Departamento de Microbiologia, Imunologia e Parasitologia - Universidade Federal de São Paulo, Escola Paulista de Medicina (UNIFESP-EPM). Rua Botucatu 862, 3° andar, 04023-062 São Paulo, SP, Brasil. (2 ) Setor de Micobactérias, Instituto Adolfo Lutz de São Paulo. Av. Dr. Arnaldo 351, 9 o andar, 01246-902 São Paulo, SP, Brasil. ), Débora de Cássia Pires GEIGER(1 (1 ) Departamento de Microbiologia, Imunologia e Parasitologia - Universidade Federal de São Paulo, Escola Paulista de Medicina (UNIFESP-EPM). Rua Botucatu 862, 3° andar, 04023-062 São Paulo, SP, Brasil. (2 ) Setor de Micobactérias, Instituto Adolfo Lutz de São Paulo. Av. Dr. Arnaldo 351, 9 o andar, 01246-902 São Paulo, SP, Brasil. ) & Sylvia Cardoso LEÃO(1 (1 ) Departamento de Microbiologia, Imunologia e Parasitologia - Universidade Federal de São Paulo, Escola Paulista de Medicina (UNIFESP-EPM). Rua Botucatu 862, 3° andar, 04023-062 São Paulo, SP, Brasil. (2 ) Setor de Micobactérias, Instituto Adolfo Lutz de São Paulo. Av. Dr. Arnaldo 351, 9 o andar, 01246-902 São Paulo, SP, Brasil. )
SUMMARY
More than 70 species of mycobacteria have been defined, and some can cause disease in humans, especially in immunocompromised patients. Species identification in most clinical laboratories is based on phenotypic characteristics and biochemical tests and final results are obtained only after two to four weeks. Quick identification methods, by reducing time for diagnosis, could expedite institution of specific treatment, increasing chances of success. PCR restriction-enzyme analysis (PRA) of the hsp65 gene was used as a rapid method for identification of 103 clinical isolates. Band patterns were interpreted by comparison with published tables and patterns available at an Internet site (http://www.hospvd.ch:8005). Concordant results of PRA and biochemical identification were obtained in 76 out of 83 isolates (91.5%). Results from 20 isolates could not be compared due to inconclusive PRA or biochemical identification. The results of this work showed that PRA could improve identification of mycobacteria in a routine setting because it is accurate, fast, and cheaper than conventional phenotypic identification.
KEYWORDS:Mycobacterium; PCR; Diagnosis; Identification
INTRODUCTION
Diagnosis of mycobacterial infections is confirmed by visualization of acid-fast bacilli by direct microscopy and/or by isolation of bacilli from biological specimens. Species identification is usually obtained, after culture in appropriate media, by tests based on phenotypic characteristics. As mycobacterial species have different drug susceptibilities, precise identification is crucial for adoption of correct drug therapy and can ultimately influence patient outcome. Different classification schemes for identification of mycobacterial species have been proposed and each laboratory adopts a preferred set of tests. M. tuberculosis, the main pathogenic mycobacterium, is usually quickly identified after isolation. For other species, final diagnosis, after isolation, can take up to 4 weeks.
In the last years, alternative rapid identification methods were developed and evaluated for clinical use, as radiometric methods associated with DNA probe hybridization5, mycolic acid analysis9, DNA sequencing11,13, and tests based on PCR10,16. Radiometric methods and DNA probes are currently used in many laboratories throughout the world, but are expensive for developing countries. Mycolic acid analysis and DNA sequencing require technical expertise and special equipment. Several authors have proposed tests based on PCR for identification of mycobacteria from cultured and non-cultured clinical isolates. These tests can be less expensive, and personnel's training is easily performed14,18. PCR is technically less demanding than direct sequencing and cheaper than identification with species-specific commercial probes.
TELENTI et al.16 described a method of mycobacterial differential diagnosis, named PRA (PCR Restriction-enzyme Analysis), based on the amplification of the hsp65 gene by PCR, followed by digestion of the amplified product with the restriction enzymes BstE II and Hae III. Twenty-nine species and subspecies were identified by analysis, on agarose gels, of restriction pattern obtained with each enzyme. The original algorithm proposed by TELENTI has been modified by TAYLOR et al.15 and DEVALLOIS et al.3. Subsequent isolated reports on variants of specific PRA patterns have increased the number of species and subspecies identified by this method1,4,8. An Internet Database (PRASITE) (http://www.hospvd.ch:8005) is available for interpretation of PRA results. Seventy-four different patterns of 40 species have so far been included in the PRASITE database.
The substitution of labor-intensive phenotypical identification for a rapid, economic and accurate test in routine laboratories is mostly desired but it would depend on careful evaluation of cost-benefits. In this work, PRA and phenotypical identification were compared in a routine setting.
MATERIAL AND METHODS
Mycobacterial isolates: For standardization of PRA in our laboratory, 15 reference strains were studied, including M. tuberculosis ATCC 14323, M. gordonae ATCC 14470, M. fortuitum ATCC 35391, and strains from Instituto Adolfo Lutz (IAL) collection, including three M. tuberculosis strains with different drug sensitivity patterns (one resistant to isoniazid, streptomycin, and pyrazinamide, one resistant to rifampicin, and one susceptible to all drugs tested), two M. kansasii, one M. bovis, one M. gordonae, one M. chelonae, two M. marinum, and two from M. avium complex (MAC) (Figure 1).
One hundred and three mycobacterial isolates from different biological specimens (sputum, blood, pus, bronchoalveolar lavage, gastric washing, lymph node biopsy, peritoneal fluid, urine, skin biopsy, cerebrospinal fluid and bone marrow aspirate), sent to IAL-SP from July to September 1998, were included in this study. Conventional and genetic identification were performed concomitantly, in different settings, and results were compared at the end of the study.
Biochemical identification: Initial presumptive distinction of M.tuberculosis from NTM (non-tuberculous mycobacteria) was obtained by direct observation of colony aspect (morphology and pigmentation) and presence of cord on Ziehl-Neelsen stained smears obtained from cultures. Cord-positive isolates were further analyzed by subcultures in Lowenstein-Jensen medium containing PNB (p-nitrobenzoic acid - 0,5 mg/ml) and TCH (2-thiophene-carboxylic acid hydrazide - 5 mg/ml) and were tested, at the same time, for susceptibility to isoniazid (INH), rifampicin (RMP), streptomycin (SM), pyrazinamide (PZA), and ethambutol (EMB) by the resistance ratio method described by COLLINS et al 2. NTM were identified by a series of conventional tests (pigmentation, growth at different temperatures, biochemical tests, tests for susceptibility to antibacterial agents), based on methods described by COLLINS et al.2, KENT & KUBICA6, and TSUKAMURA17.
Preparation of samples for PCR: One loopful of bacteria was suspended in 400 ml of TET (Tris-Cl 10mM pH 7.5, EDTA 1 mM, Triton X-100 1%) and submitted to three 10 minutes cycles of freezing and boiling. Two to ten microliters of supernatant were used for PCR.
PRA: Amplification of a 439 bp fragment from the hsp65 gene with primers Tb11 (5'-ACCAACGATGGTGTGTCCAT) and Tb12 (5'-CTTGTCGAACCGCATACCCT) was performed according to the protocol described by TELENTI et al.16. Reaction mix contained 20 mM Tris, pH 8.4, 50 mM KCl, 1.5 mM MgCl2, 10% glycerol, 200 mM dNTP, 25 pmoles of each primer and 1U Taq polymerase (CENBIOT, RS). DNA was submitted to denaturation at 95 °C for 10 min, followed by 45 cycles at 94 °C for 1 min, 60 °C for 1 min and 72 °C for 1 min and a final extension step at 72 °C for 7 min. Ten to fifteen microliters of amplified product were digested with BstE II and Hae III (Gibco/BRL) and loaded on 4% agarose gel (Gibco/BRL).
Analysis of PRA results: Scanned gel images were obtained with an EPSON ES-1000C scanner. Restriction patterns were analyzed using the Molecular Analyst program (BioRad). Fragment sizes were compared to patterns reported by DEVALLOIS et al.3. Restriction patterns were also analyzed in the PRASITE query.
RESULTS
Fifteen reference strains were submitted to PRA for standardization of this technique in the laboratory. All strains were correctly identified by comparison to published patterns and to the PRASITE database (Figure 1).
Ninety-one of 103 isolates could be identified by PRA. DNA from seven isolates did not amplify with PRA primers in two or more experiments and, in other five isolates, the fragments generated after BstE II and Hae III digestion could not be assigned to any pattern described in published algorithms or in the PRASITE database.
Biochemical identification was obtained for 89 out of 103 isolates. For the remaining 14 isolates, two did not grow, eight got contaminated in the identification tubes, and four isolates could not be identified to the species level with the tests used. The 20 isolates not properly identified by PRA and/or phenotypic identification were excluded from final analysis.
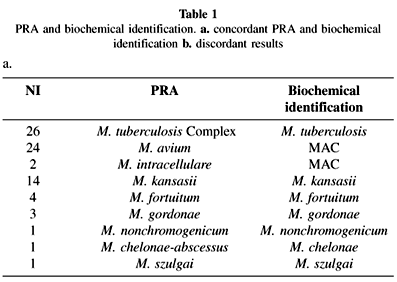
PRA and biochemical identification showed concordant results in 76 of 83 isolates (91.5%) (Table 1a). Three different M. avium PRA variants were observed, 16 presenting type I pattern, 6 type II and 2 type III, as has been previously reported by our group8. M. avium type I is the M. avium prototype pattern described in TELENTI et al.16, and corresponds to PRASITE M. avium type 2. M. avium type II corresponds to PRASITE M. avium type 1 pattern. M. avium type III has recently been included in the PRASITE database as M. avium type 3. Fourteen M. kansasii isolates presented type I PRA pattern, and 3 M. gordonae presented type III PRA pattern1,3.
In seven isolates, PRA results and phenotypic identification were discordant (Table 1b). Both identification methods were repeated, with the same discordant results.
Considering the most frequently encountered mycobacteria, M. tuberculosis and M. avium, a very good correlation between PRA and biochemical identification was observed. PRA for identification of M. tuberculosis showed 100% sensitivity, specificity of 96.4%, and positive predictive value of 92.8%. For M. avium identification the test presented 92.3% sensitivity, 98.2% specificity, and 96% of positive predictive value. Calculated kappa score for identification of M. tuberculosis was 0.945, and for M. avium, 0.915 - both values are considered almost perfect 7.
DISCUSSION
Alternative rapid methods for identification of mycobacteria can be implemented in routine settings if comparison with conventional identification methods demonstrates meaningful advantages. A comparison of PRA with commercially available molecular methods of mycobacterial identification in Brazilian isolates was recently published12, showing good correlation and cost-benefit. In our work PRA was compared to biochemical identification and the results showed that it is also a valid alternative to conventional non-molecular identification methods.
Time needed to complete PRA was less than one week, thus reducing time for final diagnosis. Considering only reagents used in both identification methods, PCR was 50% cheaper than biochemical identification. Besides that, a single person can perform PRA. Biochemical identification usually involves different persons for sterilization, preparation of media and handling of cultures. Manipulation of bacteria is minimal with PRA and significant with biochemical identification, bringing with it biosafety implications.
The main drawback in routine use of PRA is the analysis of patterns. Differences between calculated sizes of restriction fragments and published patterns have been reported3,12, and could be due to differences in running conditions, type of agarose used, or computer programs. In our experiments, fragment sizes were usually smaller than published ones. In the PRASITE query these small differences are corrected and accurate interpretation is achieved in most cases. Recently it has been proposed that a database in the GelCompar program could help in interpretation of PRA results12. Normalization and analysis steps in GelCompar could also correct for small differences in running conditions and facilitate interpretation. This program is not commonly available in most mycobacteria reference laboratories, but scanned images could be sent by email to a reference center with expertise in the use of the program and the GelCompar PRA database.
In this work, 12 isolates were not identified by PRA and 14 were not identified by phenotypical methods, giving a similar rate of failure for both methods. Seven isolates were not amplified with PRA primers. Four were identified by biochemical tests as M. kansasii (2), M. fortuitum (1) and MAC (1). The reasons for lack of amplification were not identified, but the presence of PCR inhibitors could not be ruled out. Five isolates presented novel band patterns and were not identified by PRA. Two were phenotipically identified as MAC and could correspond to allelic variants of M. avium or other members of MAC. We have recently identified two novel M. avium PRA patterns in isolates from pigs and humans8, which are different from the band patterns found in these two isolates. The remaining three isolates were not identified by biochemical tests, and could correspond to contaminants (Nocardia, Rodococcus, or other species bearing homologous hsp65 genes).
Discordant results were obtained with seven isolates. Discordant identification could result from the presence of more than one mycobacterial species in the original culture, especially if we consider those obtained from non-sterile sites. Table 1b shows that six isolates were obtained from sputum and one from blood. The discordant isolates were plated on 7H10 agar plates to obtain isolated colonies. All colonies in each plate had the same morphology and pigmentation, ruling out the occurrence of mixed cultures (data not shown). Potential pathogens were identified in six specimens (M. tuberculosis complex, M. avium, M. fortuitum, M. kansasii) by one method or the other. Clinical information of these patients was not available, and we do not know if these patients were treated, or what was their response to treatment. Another possible explanation for the discordant identification could be lack of standardization of biochemical identification and PRA patterns for uncommon or new mycobacterial species. The possibility that uncommon mycobacteria, not identified by presently used methods, could be causing disease in these patients is a matter for consideration. These seven isolates will be identified by other methods in order to elucidate this issue.
In a routine setting, quick identification of M. tuberculosis complex, M. avium, M. kansasii, and M. fortuitum is important. Treatment of diseases caused by each of these species is different, and it is important to have a rapid final diagnosis for institution of an efficient drug regimen. In our study, identification of these species by PRA could be performed by simple visualization on agarose gels. This confirms once more the usefulness of this technique, even if no computer facilities for interpretation are available. A trained person should be able to identify these most frequently encountered disease-causing mycobacteria by analysis of PRA patterns on agarose gels.
In conclusion, PRA is a valid alternative for phenotypic identification in the routine laboratory. It is fast, cheap and accurate for identification of pathogenic and potentially pathogenic mycobacteria.
RESUMO
PCR e análise de padrões de restrição do gene hsp65 (pra) para identificação de micobactérias no laboratório clínico
Mais de 70 espécies de micobactérias já foram definidas e algumas delas podem causar enfermidade em humanos, especialmente em pacientes imunocomprometidos. A identificação de espécie, na maioria dos laboratórios clínicos, se baseia em características fenotípicas e testes bioquímicos e resultados definitivos só são obtidos após duas a quatro semanas. Métodos rápidos de identificação reduzem o tempo necessário para o diagnóstico e podem antecipar a instituição do tratamento específico, aumentando as chances de sucesso. A análise de padrões de restrição do gene hsp65 amplificado por PCR (PRA) foi utilizada como método rápido de identificação em 103 isolamentos clínicos. Os padrões de bandas foram interpretados por comparação com tabelas publicadas e padrões disponíveis em um site de Internet (http://www.hospvd.ch:8005). Resultados concordantes de PRA e identificação bioquímica foram obtidos em 76 de 83 isolamentos (91,5%). Os resultados de 20 isolamentos não puderam ser comparados porque a identificação fenotípica ou por PRA foi inconclusiva. Os resultados deste trabalho mostram que PRA pode ser útil para identificação de rotina de micobactérias por ser um método acurado, rápido e mais econômico do que a identificação convencional.
ACKNOWLEDGEMENTS
We acknowledge Marcelo Burattini for careful reading of the manuscript and Rosângela Siqueira de Oliveira for technical assistance.
Financial support was obtained from FAPESP and CNPq.
Received: 02 June 2000
Accepted: 25 August 2000
Financial support: FAPESP. CFS was the recipient of a fellowship from PIBIC-CNPq
This paper received the "Premio Leal Prado Medicina Experimental", from "Pro- Reitoria de Graduação/UNIFESP", in 2000.
Correspondence to: S. C. Leão, Universidade Federal de São Paulo, Escola Paulista de Medicina, Disciplina de Microbiologia, Rua Botucatu 862, 3° andar, 04023-062 São Paulo, Brasil. Tel: (55-11) 5084-3213 Fax: (55-11) 5571-6504, e-mail: sylvia@ecb.epm.br
- 1. ALCAIDE, F.; RICHTER, I.; BERNASCONI, C. et al - Heterogeneity and clonality among isolates of Mycobacterium kansasii: implications for epidemiological and pathogenicity studies. J. clin. Microbiol., 35: 1959-1964, 1997.
- 2. COLLINS, C.H.; GRANGE, J.M. & YATES, M.D. - Tuberculosis Bacteriology Oxford, Butterworth Heinemann, 1997.
- 3. DEVALLOIS, A.; GOH, K.S. & RASTOGI, N. - Rapid identification of mycobacteria to species level by PCR-restriction fragment length polymorphism analysis of the hsp65 gene and proposition of an algorithm to differentiate 34 mycobacterial species. J. clin. Microbiol., 35: 2969-2973, 1997.
- 4. DEVALLOIS, A.; PICARDEAU, M.; PARAMASIVAN, C.N.; VINCENT, V. & RASTOGI, N. - Molecular characterization of Mycobacterium avium complex isolates giving discordant results in AccuProbe tests by PCR-restriction enzyme analysis, 16S rRNA gene sequencing, and DT1-DT6 PCR. J. clin. Microbiol., 35: 2767-2772, 1997.
- 5. ELLNER, P.D.; KIEHN, T.E.; CAMMARATA, R. & HOSMER, M. - Rapid detection and identification of pathogenic mycobacteria by combining radiometric and nucleic acid probe methods. J. clin. Microbiol., 26: 1349-1352, 1988.
- 6. KENT, P.T. & KUBICA, G.P. - Public health mycobacteriology: a guide for the level III laboratory Atlanta, US Department of Health and Human Services; Centers for Disease Control, 1985.
- 7. LANDIS, J.R. & KOCH, G.G. - An application of hierarchical kappa-type statistics in the assessment of majority agreement among multiple observers. Biometrics, 33: 363-374, 1977.
- 8. LEĂO, S.C.; BRIONES, M.R.S.; SIRCILI, M.P. et al - Identification of two novel Mycobacterium avium allelic variants in pig and human isolates from Brazil by PCR-restriction enzyme analysis. J. clin. Microbiol., 37: 2592-2597, 1999.
- 9. LEITE, C.Q.; DE SOUZA, C.W. & LEITE, S.R. - Identification of mycobacteria by thin layer chromatographic analysis of mycolic acids and conventional biochemical method: four years of experience. Mem. Inst. Oswaldo Cruz, 93: 801-805, 1998.
- 10. NINET, B.; ROHNER, P.; METRAL, C. & AUCKENTHALER, R. - Assessment of use of the COBAS AMPLICOR system with BACTEC 12B cultures for rapid detection of frequently identified mycobacteria. J. clin. Microbiol., 37: 782-784, 1999.
- 11. PAI, S.; ESEN, N.; PAN, X. & MUSSER, J.M. - Routine rapid Mycobacterium species assignment based on species-specific allelic variation in the 65-kilodalton heat shock protein gene (hsp65). Arch. Path. Lab. Med., 121: 859-864, 1997.
- 12. ROCHA, A.S.; LEITE, C.C.; TORRES, H.M. et al. - Use of PCR-restriction fragment length polymorphism analysis of the hsp65 gene for rapid identification of mycobacteria in Brazil. J. microbiol. Meth., 37: 223-229, 1999.
- 13. SPRINGER, B.; STOCKMAN, L.; TESCHNER, K.; ROBERTS, G.D. & BÖTTGER, E.C. - Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J. clin. Microbiol., 34: 296-303, 1996.
- 14. TAKEWAKI, S.; OKUZUMI, K.; ISHIKO, H. et al - Genus-specific polymerase chain reaction for the mycobacterial dnaJ gene and species-specific oligonucleotide probes. J. clin. Microbiol., 31: 446-450, 1993.
- 15. TAYLOR, T.B.; PATTERSON, C.; HALE, Y. & SAFRANEK, W.W. - Routine use of PCR-restriction fragment length polymorphism analysis for identification of mycobacteria growing in liquid media. J. clin. Microbiol., 35: 79-85, 1997.
- 16. TELENTI, A.; MARCHESI, F.; BALZ, M. et al - Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. clin. Microbiol., 31: 175-178, 1993.
- 17. TSUKAMURA, M. - Identification of mycobacteria Obu, National Chubu Hospital, 1984.
- 18. VANEECHOUTTE, M.; DE BEENHOUWER, H.; CLAEYS, G. et al - Identification of Mycobacterum species by using amplified ribosomal DNA restriction analysis. J. clin. Microbiol., 31: 2061-2065, 1993.
Publication Dates
-
Publication in this collection
09 Mar 2001 -
Date of issue
Feb 2001
History
-
Received
02 June 2000 -
Accepted
25 Aug 2000