Abstracts
Trypanosoma rangeli is non pathogenic for humans but of important medical and epidemiological interest because it shares vertebrate hosts, insect vectors, reservoirs and geographic areas with T. cruzi, the etiological agent of Chagas disease. Therefore, in this work, we set up two PCR reactions, TcH2AF/R and TrFR2, to distinguish T. cruzi from T. rangeli in mixed infections of vectors based on amplification of the histone H2A/SIRE and the small nucleolar RNA Cl1 genes, respectively. Both PCRs were able to appropriately detect all T. cruzi or T. rangeli experimentally infected-triatomines, as well as the S35/S36 PCR which amplifies the variable region of minicircle kDNA of T. cruzi. In mixed infections, whereas T. cruzi DNA was amplified in 100% of samples with TcH2AF/R and S35/S36 PCRs, T. rangeli was detected in 71% with TrF/R2 and in 6% with S35/S36. In a group of Rhodnius colombiensis collected from Coyaima (Colombia), T. cruzi was identified in 100% with both PCRs and T. rangeli in 14% with TrF/R2 and 10% with S35/S36 PCR. These results show that TcH2AF/R and TrF/R2 PCRs which are capable of recognizing all T. cruzi and T. rangeli strains and lineages could be useful for diagnosis as well as for epidemiological field studies of T. cruzi and T. rangeli vector infections.
Trypanosoma cruzi; Trypanosoma rangeli; Rhodnius prolixus; Rhodnius colombiensis; PCR; Histone H2A; SIRE; sno-RNA -Cl1
Embora o Trypanosoma rangeli não seja patogênico para o homem, sua importância médica e epidemiológica reside no fato de compartilhar vetores, reservatórios e áreas geográficas com o Trypanosoma cruzi, agente causal da Doença de Chagas. Neste estudo, para distinguir T. cruzi de T. rangeli em vetores com infecções mistas, se utilizaram duas amplificações de PCR; TcH2AF/R para o gen da histona H2A/SIRE e TrFR2, para um gen repetitivo de ARN nucleolar Cl1 (sno-RNA-Cl1). Assim como a PCR S35/S36, ambas as reações foram capazes de detectar corretamente a presença de T. cruzi ou T. rangeli em triatomíneos infectados experimentalmente. Nas infecções mistas, o ADN de T. cruzi foi amplificado em 100% das amostras quando se utilizaram TcH2AF/R e S35/S36, enquanto T. rangeli foi detectado em 71% delas com os iniciadores TrF/R2 e em 6%, com S35/S36. Adicionalmente, em um grupo de Rhodnius colombiensis coletados na região de Coyaima (Tolima), T. cruzi foi identificado em 100% com ambas PCRs e T. rangeli em 14% delas com os iniciadores TrF/R2 e em 10%, com S35/S36. Estes resultados mostram que as reações de PCR TcH2AF/R e TrF/R2, capazes de reconhecer todas as cepas e linhagens de T. cruzi e T. rangeli, podem ser úteis no diagnóstico e também nos estudos epidemiológicos do campo com vetores infectados pelo T. cruzi e T. rangeli.
TRYPANOSOMIASIS
Detection of Trypanosoma cruzi and Trypanosoma rangeli infection in triatomine vectors by amplification of the histone H2A/SIRE and the sno-RNA-C11 genes
Detecção da infecção por Trypanosoma cruzi e Trypanosoma rangeli em vetores triatomíneos através da amplificação dos gens de histona H2A/SIRE e sno-RNA-C11
Paula Ximena PaviaI; Gustavo Adolfo VallejoII; Marleny MontillaIII; Rubén Santiago NichollsIII; Concepción Judith PuertaI
ILaboratorio de Parasitología Molecular, Departamento de Microbiología, Facultad de Ciencias, Pontificia Universidad Javeriana, Carrera 7 No. 43-82, Edificio 50, Laboratorio 113, Bogotá, Colombia
IILaboratorio de Investigaciones en Parasitología Tropical, Facultad de Ciencias, Universidad del Tolima, A.A. No. 546, Ibagué, Colombia
IIILaboratorio de Parasitología, Instituto Nacional de Salud, Avenida- Calle 26 No. 51-60, Bogotá, Colombia
Correspondence to Correspondence to: Concepción Judith Puerta Tel: +57-3208320 ext. 4024; Fax: +57-3 208320 ext. 4021 E-mail: cpuerta@javeriana.edu.co
SUMMARY
Trypanosoma rangeli is non pathogenic for humans but of important medical and epidemiological interest because it shares vertebrate hosts, insect vectors, reservoirs and geographic areas with T. cruzi, the etiological agent of Chagas disease. Therefore, in this work, we set up two PCR reactions, TcH2AF/R and TrFR2, to distinguish T. cruzi from T. rangeli in mixed infections of vectors based on amplification of the histone H2A/SIRE and the small nucleolar RNA Cl1 genes, respectively. Both PCRs were able to appropriately detect all T. cruzi or T. rangeli experimentally infected-triatomines, as well as the S35/S36 PCR which amplifies the variable region of minicircle kDNA of T. cruzi. In mixed infections, whereas T. cruzi DNA was amplified in 100% of samples with TcH2AF/R and S35/S36 PCRs, T. rangeli was detected in 71% with TrF/R2 and in 6% with S35/S36. In a group of Rhodnius colombiensis collected from Coyaima (Colombia), T. cruzi was identified in 100% with both PCRs and T. rangeli in 14% with TrF/R2 and 10% with S35/S36 PCR. These results show that TcH2AF/R and TrF/R2 PCRs which are capable of recognizing all T. cruzi and T. rangeli strains and lineages could be useful for diagnosis as well as for epidemiological field studies of T. cruzi and T. rangeli vector infections.
Keywords:Trypanosoma cruzi; Trypanosoma rangeli; Rhodnius prolixus; Rhodnius colombiensis; PCR, Histone H2A; SIRE; sno-RNA -Cl1.
RESUMO
Embora o Trypanosoma rangeli não seja patogênico para o homem, sua importância médica e epidemiológica reside no fato de compartilhar vetores, reservatórios e áreas geográficas com o Trypanosoma cruzi, agente causal da Doença de Chagas. Neste estudo, para distinguir T. cruzi de T. rangeli em vetores com infecções mistas, se utilizaram duas amplificações de PCR; TcH2AF/R para o gen da histona H2A/SIRE e TrFR2, para um gen repetitivo de ARN nucleolar Cl1 (sno-RNA-Cl1). Assim como a PCR S35/S36, ambas as reações foram capazes de detectar corretamente a presença de T. cruzi ou T. rangeli em triatomíneos infectados experimentalmente. Nas infecções mistas, o ADN de T. cruzi foi amplificado em 100% das amostras quando se utilizaram TcH2AF/R e S35/S36, enquanto T. rangeli foi detectado em 71% delas com os iniciadores TrF/R2 e em 6%, com S35/S36. Adicionalmente, em um grupo de Rhodnius colombiensis coletados na região de Coyaima (Tolima), T. cruzi foi identificado em 100% com ambas PCRs e T. rangeli em 14% delas com os iniciadores TrF/R2 e em 10%, com S35/S36. Estes resultados mostram que as reações de PCR TcH2AF/R e TrF/R2, capazes de reconhecer todas as cepas e linhagens de T. cruzi e T. rangeli, podem ser úteis no diagnóstico e também nos estudos epidemiológicos do campo com vetores infectados pelo T. cruzi e T. rangeli.
INTRODUCTION
Chagas disease, caused by the flagellate parasite Trypanosoma cruzi, affects fifteen countries throughout Latin America. The number of new cases has been estimated at 200,000 per year and about 21,000 chagasic patients die each year from the disease17,42. In Colombia, Chagas disease is a major public health problem. It is estimated that 5% of the population is at a high risk of being infected and that approximately 700,000 people are currently infected17. Chagas disease is transmitted to humans by bloodsucking triatomine bugs, from which 23 species have been reported in Colombia. The most important vector species that circulate in the domestic cycle in this country are Rhodnius prolixus, Triatoma dimidiata, Triatoma maculata, and Triatoma venosa whereas in the sylvatic regions Rhodnius pallescens and Rhodnius colombiensis are the prevalent ones12,15.
Based on phenotypic and genotypic characters, T. cruzi has been divided into two principal lineages: T. cruzi I and T. cruzi II1. In addition, T. cruzi II is divided into five subgroups, named IIa-e2. While T. cruzi I, associated with opossums and an arboreal ecology, predominates from the Amazon basin northwards, T. cruzi II is associated with armadillos and a terrestrial ecology and predominates in southern cone countries of South America43.
On the other hand, although Trypanosoma rangeli infection in humans is harmless, this parasite is a serious concern for the epidemiology and diagnosis of Chagas disease due to its morphological similarity and immunological cross-reactivity with T. cruzi14. Moreover, these trypanosomes are sympatric and share triatomine insects as well as vertebrate hosts, allowing the occurrence of mixed infections7. Colombia is one of the countries in which T. cruzi shares vectors and reservoirs with T. rangeli7,13,14. Recently, two important epidemiological groups of T. rangeli have been described: KP1(-) strains, associated with the adaptive line of Rhodnius, represented by R. colombiensis, R. pallescens, and R. ecuadoriensis, and KP1(+) strains, associated with R. prolixus30-32. These two groups have been defined on the basis of independent mitochondrial (minicircle profile dimorphism obtained by kDNA PCR amplification) and nuclear (mini-exon PCR amplification) markers.
Due to direct microscopic detection of trypanosomes, the traditional method for assessment of infection in vectors is not able to distinguish T. cruzi from T. rangeli infection, several polymerase chain reaction techniques have been developed3,4,5,6,9,13,19,28,33,36,40,41. However, current PCR assays used for mixed infection detection show some disadvantages such as the amplification of bands of similar size both in T. cruzi and T. rangeli27,28, the amplification of polymorphic fragments11,19, and bias to T. cruzi in the case of mixed T. cruzi and T. rangeli infection9,33,36. Therefore, it is necessary to develop new techniques as better options to specifically identify each of these parasite species in mixed infections. In this study we aimed to determine the capacity of two previously standardized PCR tests to detect specifically T. rangeli and T. cruzi based on the small nucleolar RNA- C11 (sno-RNA-C11) gene and the SIRE (short interspersed repetitive element) sequence inserted into the histone H2A gene, respectively18,20. Our results indicate that the use of both PCR tests allow a specific identification of T. cruzi and T. rangeli in experimental and natural triatomine infection.
MATERIALS AND METHODS
Parasites: Artificial and experimental infections were performed with the Colombian strains MHOM/CO/01/DA and IRHO/CO/85/MTA, belonging to T. cruzi I, and the Colombian KP1(+) strain IRHO/CO/86/CH, of T. rangeli. These strains were provided by Laboratorio de Parasitología, Instituto Nacional de Salud, (INS) (Bogotá, Colombia). Both parasites species were characterized by isoenzymes24 and PCR using miniexon and minicircle sequences as targets5,31,33. The bulk parasite mass was cultivated in REI modified liquid medium, supplemented with 2% FCS and 100 µg/mL of gentamicin, and incubated at 24 ºC.
Triatomines: Experimental infections were carried out using non-infected fifth stage nymphs of R. prolixus. Fourteen specimens were fed on mice previously infected with 1 x 106 flagellates/mL of T. cruzi. Twenty-two insects were infected with T. rangeli by intrafemoral inoculation of 1 x 105 parasites/mL. Seventeen triatomines were fed on mice inoculated intraperitoneally with 1 x 106 of both trypanosomes per mL. Forty non-infected vectors were used as controls and distributed equally among each group. On the other hand, twenty-nine specimens of R. colombiensis were collected from Attalea butyracea palm trees in Coyaima, Department of Tolima (Colombia) and 21 T. maculata specimens were collected from the Department of Bolivar (Colombia). Each experimentally infected triatomine was dissected 15 and 45 days post-infection to obtain the intestinal tract, feces, hemolymph and salivary glands respectively after exposure to UV light for one hour. Samples were homogenized in 240 µL of phosphate buffered saline (PBS). A 40 µL aliquot was examined by direct microscope observation (DMO) and Giemsa stained. The DNA from the rest of the sample was extracted three times with phenol-chloroform-isoamylic alcohol (25:24:1), followed by ethanol precipitation33. Finally, DNA obtained was amplified with the different PCR methods. Field-collected triatomines were treated and analyzed as described above. To simulate vector infection, the digestive tracts of non-infected adult triatomines (R. prolixus) were removed (100 µL) and mixed with 100 µL of 1 to 104 epimastigote forms of T. cruzi or 2 x 106 to 12 x 106 epimastigote forms of T. rangeli. Then, DNA was extracted and amplified with different PCR tests.
PCR conditions: For T. cruzi detection the following primers were used: TcH2AF (5'-GAGAGTGATCGTGGGAGAGC-3'), and TcH2AR (5'-AGTGGCAGACTTTGG GGTC-3'). These primers amplify a 234 bp fragment present in the 3' non-encoding region of the 1.2 kb unit encoding for histone H2A from T. cruzi (GenBank accession number X67287), corresponding to the 16-248 nucleotides of SIRE sequence20,21. PCR reactions were performed in a 25 µL final volume, containing: 5 µL of DNA from triatomines (diluted 1:5 or 1:10), 1X reaction buffer (10 mM Tris-HCl, pH 8.5, 500 mM KCl), 1.25 U/µL of Taq DNA polymerase, 1.5 mM of MgCl2, 200 µM of deoxynucleoside triphosphate (dNTP) mixture, and 20 pmol of each primer. The reaction was carried out on a MJ Research PTC-100 DNA thermal cycler, using the following profile: 95 ºC/5 min, 15 cycles of 95 ºC/30 s, 72 ºC/1 min, and 30 cycles of 95 ºC/30 s, 65 ºC/30 s and 72 ºC/30 s, and a final incubation of 72 ºC for five min. 15 µL of the reaction products were electrophoresed in 1.5% agarose gels and stained with ethidium bromide. For T. rangeli detection, we used a modified version of a TrF/R2 PCR assay targeted to the sno-RNA-Cl1 genes (GenBank accession number AY028385) which amplifies a 620 bp fragment18. PCR reactions were made in a 25 µL final volume, containing: 5 µL of DNA from triatomines (diluted 1:5 or 1:10), 1X reaction buffer (100 mM Tris-HCl, pH 8.5, 500 mM KCl), 1.25 U/µL of Taq DNA polymerase, 1.5 mM of MgCl2, 200 µM of dNTP mixture, and five pmol of each primer. The reaction was carried out on a MJ Research PTC-100 DNA thermal cycler, using the following profile: 95 ºC/5 min, 15 cycles of 95 ºC/30 s, 63 ºC/1 min and 72 ºC/30 s and 20 cycles of 95 ºC/30 s, 60 ºC/1 min, 72 ºC/30 s, and a final incubation of 72 ºC for 5 min. 15 µL of the reaction products were electrophoresed in 1% agarose gels and stained with ethidium bromide. In addition, all DNA samples were amplified with the S35 (5'-AAATAATGTACGGGTGGAGATGCATGA-3'), and S36 (5'-GGGTTCGATTGGGGTTGGTGT-3') primers based on conserved regions of the minicircles from kDNA from T. cruzi29 (GenBank accession number X04680). These primers amplify two fragments of 300 and 450 bp in T. rangeli, besides a 330 bp band amplified in T. cruzi. PCR reactions were performed according to previously described33.
RESULTS
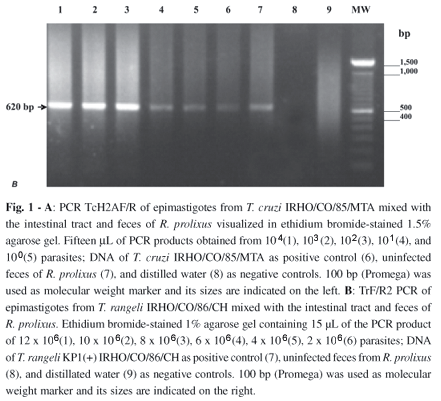
Artificial infections: To asses the sensitivity of TcH2AF/R and TrF/R2 PCRs, an artificial infection was simulated by mixing T. cruzi or T. rangeli with the intestinal tract and feces from non-infected triatomines. For T. cruzi, the appropriately sized amplification product was obtained from a single parasite (Fig. 1A, lane 5). For T. rangeli, the assay detected DNA from 2 x 106 parasites (Fig. 1B, lane 6).
Experimental infection of R. prolixus with T. cruzi: Parasites were detected 15 days post-infection in 100% (14/14) and 78% (11/14) of insects by DMO and Giemsa staining, respectively. TcH2AF/R and S35/S36 PCR reactions showed the presence of T. cruzi in 100% of insects (14/14) with the expected PCR products (Table 1). The kappa index between both PCR reactions was 1.0 (95% Confidence Interval), (95% CI: 0.95-1.0), interpreted as a perfect concordance25. The group of nine non-infected vectors all tested negative.
Experimental infection of R. prolixus with T. rangeli: The presence of parasites in the haemolymph 15 days post-infection, was 100% (22/22) determined by DMO and Giemsa staining. T. rangeli was detected in 100% (22/22) in the haemolymph with both PCR tests showing the expected amplification products (Table 2). The kappa index between both tests was 1.0 (95% CI: 0.95-1.0), interpreted as a perfect concordance25. On the other hand, non-infected triatomines were tested negative. After 45 days post-infection, 21 out of 22 T. rangeli infected vectors and three controls were decontaminated and dissected obtaining the salivary glands. Parasites were observed in 90% (19/21) and 67% (14/21) by DMO and Giemsa staining, respectively. TrF/R2 positive PCR reaction for T. rangeli was found in 33% (7/21) of vectors while 48% of insects were positive by the S35/S36 PCR (10/21) (Table 3). The kappa index between both PCR reactions was 0.76, interpreted as a good concordance (95% CI: 0.52-0.90)25. It is important to bear in mind that positive triatomine bugs by PCR were also positive with the conventional tests. The group of non-infected vectors tested negative.
Experimental infection of R. prolixus with T. cruzi and T. rangeli: The presence of parasites in the intestinal tract and feces 15 days post-infection was detected by ODM and Giemsa staining in 100% (17/17) and 76% (13/17) of cases, respectively. T. cruzi was observed in 100% (17/17) with both PCR tests (Table 4). T. rangeli was detected with TrF/R2 PCR in 71% of vectors (12/17) (Fig. 4C) and 6% (1/17) of insects with S35/S36 PCR. The group of five non-infected vectors tested negative.
Detection of T. cruzi and T. rangeli in triatomine field samples: Having studied the detection capacity of TcH2AF/R and TrF/R2 PCRs in vectors, the next step was to evaluate the T. cruzi and T. rangeli detection in naturally infected triatomines. Two groups of field triatomines, R. colombiensis and T. maculata were collected and their intestinal tract and feces were analyzed. In R. colombiensis, trypanosomes were detected in the intestinal tract and feces of 83% (23/29) by DMO and in 86% (25/29) by Giemsa staining, whereas in salivary glands no parasites were observed.
The presence of T. cruzi in the intestinal tract and feces was detected in 100% (29/29) of insects with both PCR tests (Table 5), while T. rangeli was observed in intestinal tract and feces with TrF/R2 PCR in 14% (4/29) (Table 5) and 10% (3/29) with S35/S36 PCR. Remarkably, in eight salivary glands studied, this parasite was not observed with the tests performed, even when three out of eight specimens were positive in feces with both conventional and PCR techniques. All tests yielded negative results for both parasites in 21 T. maculata specimens.
DISCUSSION
The detection of T. rangeli and T. cruzi in naturally infected vectors has been subject to study because in intestinal infections both species of flagellates can be found and the distinction between them is difficult35.
Bearing this in mind, in this work we evaluated the use of TcH2AF/R20 and TrF/R2 PCRs18 to identify these trypanosomes by comparing them with conventional techniques and with the S35/S36 PCR, the most sensitive PCR described for T. cruzi3,4,40,41 in the two last decades. Despite the S35/S36 PCR being described as a specific tool for the amplification of kDNA from T. cruzi at the beginning8,29, sequencing of kDNA minicircles of T. rangeli found a high degree of homology among conserved regions between the two species. Therefore, these primers anneal and amplify kDNA from both trypanosomes33,34.
TcH2AF/R amplification is a highly specific test for detecting DNA from T. cruzi I and T. cruzi II strains which yields an amplification product of the same size in both T. cruzi groups. In addition, this PCR does not amplify T. rangeli DNA as well as DNA from other trypanosomatids, humans, mice or vectors20. In this study, we could detect the presence of T. cruzi DNA by PCR in artificial infections from a single parasite. Also, we detected T. cruzi in intestinal tract and feces in 100% of the insects infected experimental and naturally, results that were similar to those obtained with the S35/S36 PCR. These results together with the finding that TcH2AF/R PCR can detect an equivalent to 1/200th of T. cruzi cells20, suggest this PCR can be considered as one of the main techniques which display a great power of detection in insect vectors among those that use nuclear DNA as target. The high sensitivity of this PCR might be explained because this PCR amplifies a fragment corresponding to the 16-248 nucleotides of SIRE, a short interspersed repetitive element, present in T. cruzi21,38,39. SIRE, a sequence of 428 bp is repeated about 1500 to 3000 times per genome depending on the parasite strain but to date it has not been reported in T. rangeli23,38,39. However, recent studies show the presence of a retrotransposon inserted in nucleotide 182 of SIRE in T. cruzi16,37 and Trypanosoma brucei16, rising to a VIPER (Vestigial interposed retroelement) of 4480 bp in length16. This element, a tyrosine recombinase retroelement, is composed by three open reading frames (ORF) flanked by the first 182 bp of SIRE in the 5' region and by the last 226 bp of SIRE in the 3' region. Therefore, in the case that VIPER would be present in the genome of T. rangeli this retroelement should not amplify with the TcH2AF/R primers because SIRE is blocked at the nucleotide 182 by the specific 4072 bp from VIPER. Interestingly, ELIAS et al. (2003)10 and SCHIJMAN et al. (2004)26 reported a nested PCR based on SIRE which is able of detecting parasite DNA in the heart tissue of chronic chagasic patients.
On the other hand, the TrF/R2 PCR, based on the repetitive genes encoding for the sno-RNA-Cl1, is a specific test that amplify a product of the same size in both T. rangeli KP1(+) and KP1(-) strains and does not present any amplification signal either with the DNA from both T. cruzi and DNA from human, mouse or even other trypanosomatids18. In this work, with this PCR T. rangeli was detected in a specific way in artificial infections from 2 x 106 parasites. In addition, in experimental infections this parasite was detected in the hemolymph of 100% of the insects using both TrF/R2 and S35/S36 PCR, while in salivary glands the parasite was detected in only 33% with TrF/R2 and in 48% with S35/S36. These results are evidence of a lower sensitivity for both PCR tests to detect T. rangeli in salivary glands in comparison with conventional methods, fact that could be due to the presence of PCR inhibitors. Otherwise, it is important to mention that detection of single infections in vectors using TrF/R2 is lower compared to the S35/S36 PCR probably due to the higher copy number of the S35/S36 target than TrF/R2 target.
However, in mixed infections with both parasites, T. rangeli was detected in intestinal tract and feces in 71% of insects with TrF/R2 primers and in 6% with S35/S36 PCR. In the same way, the analyses of intestinal tract and feces of 29 R. colombiensis showed a T. cruzi amplification profile that overlapped the presence of T. rangeli.
These results show that despite the minor copy number of snoRNA-Cl1 genes than minicircles sequences, the TrF/R2 PCR does not present primer interference with T. cruzi genome renders this PCR a very useful to detect the presence of T. rangeli in co-infected insects. Besides, this findings are in accordance with those of VALLEJO et al. (1999)33 and VARGAS et al. (2000)36 who observed that the amplification of T. cruzi with the S35/S36 primers are dominant in most mixed infections, probably because the minicircles annealing sites of T. cruzi are present in greater quantity and could compete for the annealing of primers generating a typical T. cruzi profile that overlaps the presence of T. rangeli. It is important to highlight that eight salivary glands analyzed from the 29 R. colombiensis tested negative by PCR as well as by conventional methods, suggesting that at the time of the analysis, the parasite had not yet invaded the salivary glands of insects. Besides, the negativity for T. cruzi and T. rangeli of the 21 specimens of T. maculata collected in Bolívar with both TcH2AF/R and TrF/R2 PCR's as well with the conventional techniques and the S35/S36 PCR, confirms the specificity of the TcH2AF/R and TrF/R2 PCR tests. Of special interest is the fact that the presence of T. cruzi DNA does not interfere with the amplification of the T. rangeli DNA despite the smaller copy number of the snoRNA-Cl1 genes in comparison to the TcH2AF/R targets. In addition, these two PCRs amplify different nuclear targets whose amplification product have sizes easily differentiated in a single agarose gel of 1.5%.
Among different research groups, different PCR detecting systems have been reported for detecting T. cruzi and T. rangeli in the intestinal content from experimental and naturally infected vectors. In this sense, DORN et al. (1999)9 showed that the TC1/TC2 minicircle targed PCR assay was biased to T. cruzi since T. rangeli must constitute at least 75% of the sample in the presence of T. cruzi for detection by the PCR.
Later on, VARGAS et al. (2000)36 used three PCR systems: S35/S36 (kDNA), D72/D75/RG3 (Variable domain of the ribosomal subunit (LSU) of trypanosomatids), and R1/R2 (specific repetitive element P542), found in 50% of R. colombiensis examined (6/12), T. rangeli was masked by the amplification of T. cruzi using the S35/S36 primers, concluding that this PCR does not allow the diagnosis of mixed infections in most of the insects. Afterwards, RAMÍREZ et al. (2002)22 detected T. cruzi and T. rangeli in an endemic area of Brazil using five PCR systems: S35/S36, D72/D75/RG3, R1/R2, D71/D72 (D7 domain of LSU gene from T. cruzi) and multiplex PCR for intergenic regions in the mini-exon gene. The use of the S35/S36 primers in cultured parasites showed that in 11% of the samples (2/18), T. cruzi profiles masked T. rangeli. With this, we can conclude that those primers do not allow the detection of mixed infections. A duplex PCR assay based on telomeres sequences have been developed to determine the presence of both parasites, nevertheless, since the number of samples analyzed was reduced to a single triatomine bug6, more studies are needy in order to asses the lack of interference or competition between T. cruzi and T. rangeli DNA telomeric sequences in field samples.
In order to avoid the interaction between the DNA of both parasites, it is recommended to use two independent PCR systems to detect mixed infections by T. rangeli and T. cruzi. The use of PCR reactions based on genes encoding for H2A/SIRE (TcH2AF/R) and sno-RNA-C11 (TrF/R2) to detect T. cruzi and T. rangeli, as described in the present work, provide the conditions of reliable tests to be implemented in detection of mixed infections in naturally infected triatomines since they yield non-polymorphic amplification products of different size in all T. cruzi and T. rangeli strains and lineages, and are capable of detecting both parasites in natural mixed infections. Therefore, these techniques might fulfil the conditions to become powerful tools for the study and understanding the epidemiology of Chagas disease. At present these PCR techniques are being tested as an alternative diagnostic method in humans.
ACKNOWLEDGMENTS
We would like to thank to Dr. Pedro Blanco, University of Sucre, who kindly provided the Triatoma maculata specimen. To Dr. Marcela Mercado, for her helpful assistance on the statistical analysis, and people from the Molecular Parasitology, Department of Microbiology, Pontificial Javeriana University. To the Parasitology lab of the Colombian National Institute of Health, and to the Tropical Parasitology Reaserch Group, University of Tolima, for their invaluable financial and logistic assistance.
Received: 27 April 2006
Accepted: 15 August 2006
- 1. ANONYMOUS - Recommendations from a satellite meeting. Mem. Inst. Oswaldo Cruz, 94 (suppl. 1): 429-432, 1999.
- 2. BRISSE, S.; VERHOEF, J. & TIBAYRENC, M. - Characterisation of large and small subunit rRNA and mini-exon genes further supports the distinction of six Trypanosoma cruzi lineages. Int. J. Parasit., 31: 1218-1226, 2001.
- 3. BRITTO, C.; SILVEIRA, C.; CARDOSO, M.A. et al - Parasite persistence in treated chagasic patients revealed by xenodiagnosis and polymerase chain reaction. Mem. Inst. Oswaldo Cruz, 96: 823-826, 2001.
- 4. BRITTO, C.; CARDOSO, M.A.; VANNI, C.M. et al - Polymerase chain reaction detection of Trypanosoma cruzi in human blood samples as a tool for diagnosis and treatment evaluation. Parasitology, 110: 241-247, 1995.
- 5. CAMPBELL, D.; GONZALEZ, C.; JARAMILLO, C. et al - Resumen del taller sobre el uso de la reacción en cadena de la polimerasa (PCR) para distinguir entre Trypanosoma cruzi y Trypanosoma rangeli Biomédica, 2: 94-100, 1993.
- 6. CHIURILLO, M.A.; CRISANTE, G.; ROJAS, A. et al - Detection of Trypanosoma cruzi and Trypanosoma rangeli infection by duplex PCR assay based on telomeric sequences. Clin. Diagn. Lab. Immunol., 10: 775-779, 2003.
- 7. D' ALESSANDRO, A. & SARAVIA, N. - Trypanosoma rangeli In: GILLES, H.M., ed. Protozoal diseases London, Arnold Publisher, 1999. v. 1, p. 398-412.
- 8. DEGRAVE, W.; FRAGOSO, S.P.; BRITTO, C. et al - Peculiar sequence organization of kinetoplast DNA minicircles from Trypanosoma cruzi Molec. Biochem. Parasit., 27: 63-70, 1988.
- 9. DORN, P.L.; ENGELKE, D.; RODAS, A. et al - Utility of polymerase chain reaction in detection of Trypanosoma cruzi in Guatemalan Chagas disease vectors. Amer. J. trop. Med. Hyg., 60: 740-745, 1999.
- 10. ELIAS, F.E.; VIGLIANO, C.A.; LAGUENS, R.P.; LEVIN, M.J. & BEREK, C. - Analysis of the presence of Trypanosoma cruzi in the heart tissue of three patients with chronic Chagas' heart disease. Amer. J. trop. Med. Hyg., 68: 242-247, 2003.
- 11. GRISARD, E.C.; CAMPBELL, D.A. & ROMANHA, A.J. - Mini-exon gene sequence polymorphism among Trypanosoma rangeli strains isolated from distinct geographical regions. Parasitology, 118: 375-382, 1999.
- 12. GUHL, F.; ANGULO V.M.; RESTREPO, M.; NICHOLLS, S. & MONTOYA, R. - Estado del arte de la enfermedad de Chagas en Colombia y estrategias de control. Biomédica, 23 (suppl. 1): 31-33, 2003.
- 13. GUHL, F.; JARAMILLO, C.; CARRANZA, J.C. & VALLEJO, G.A. - Molecular characterization and diagnosis of Trypanosoma cruzi and T. rangeli Arch. med. Res., 33: 362-370, 2002.
- 14. GUHL, F. & VALLEJO, G.A. - Trypanosoma (Herpetosoma) rangeli Tejera, 1920: an updated review. Mem. Inst. Oswaldo Cruz, 98: 435-442, 2003.
- 15. JARAMILLO, C.; MONTAÑA, M.F.; CASTRO, L.R.; VALLEJO, G.A. & GUHL, F. - Differentiation and genetic analysis of Rhodnius prolixus and Rhodnius colombiensis by rDNA and RAPD amplification. Mem. Inst. Oswaldo Cruz, 96: 1043-1048, 2001.
- 16. LORENZI, H.A.; ROBLEDO, G. & LEVIN, M.J. - The VIPER elements of trypanosomes constitute a novel group of tyrosine recombinase-enconding retrotransposons. Molec. Biochem. Parasit., 145: 184-194, 2006.
- 17. MONCAYO, A. - Chagas disease: current epidemiological trends after the interruption of vectorial and transfusional transmission in the Southern Cone countries. Mem. Inst. Oswaldo Cruz, 98: 577-591, 2003.
- 18. MORALES, L.; ROMERO, I.; DIEZ, H. et al - Characterization of a candidate Trypanosoma rangeli small nucleolar RNA gene and its application in a PCR-based parasite detection. Exp. Parasit., 102: 72-80, 2002.
- 19. MURTHY, V.K.; DIBBERN, K.M. & CAMBPELL, D.A. - PCR amplification of mini-exon genes differentiates Trypanosoma cruzi from Trypanosoma rangeli. Molec. Cell. Probes, 6: 237-243, 1992.
- 20. PAVÍA, P.; CUERVO, C.; MONTILLA, M.; NICHOLLS, S. & PUERTA, C. - Diseño y estandarización de una prueba de PCR para la detección específica de Trypanosoma cruzi Infectio, 7: 129-136, 2003.
- 21. PUERTA, C.; MARTIN, J.; ALONSO, C. & LOPEZ, M.C. - Isolation and characterization of the gene encoding histone H2A from Trypanosoma cruzi. Molec. Biochem. Parasit., 64: 1-10, 1994.
- 22. RAMÍREZ, L.E.; LAGES-SILVA, E.; ALVARENGA-FRANCO, F. et al - High prevalence of Trypanosoma rangeli and Trypanosoma cruzi in opossums and triatomids in a formerly-endemic area of Chagas disease in Southeast Brazil. Acta trop., 84: 189-198, 2002.
- 23. REQUENA, J.M.; JIMENEZ-RUIZ, A.; SOTO, M.; LOPEZ, M.C. & ALONSO, C. - Characterization of a highly repeated interspersed DNA sequence of Trypanosoma cruzi: its potential use in diagnosis and strain classification. Molec. Biochem. Parasit., 51: 271-280, 1992.
- 24. RODRIGUEZ, P.; MONTILLA, M.; NICHOLLS, S.; ZARANTE, I. & PUERTA, C. - Isoenzymatic characterization of Colombian strains of Trypanosoma cruzi Mem. Inst. Oswaldo Cruz, 93: 739-740, 1998.
- 25. SACKETT, D.; HAYNES, R.; GUYATT, G. & TUGWELL, P. - Selección de pruebas diagnosticas. In: Epidemiologia clínica ciencia básica para la medicina clínica. Mexico, Editorial Médica Panamericana, 1998. p. 64-67.
- 26. SCHIJMAN, A.G.; VIGLIANO, C.A.; VIOTTI, R.J. et al. - Trypanosoma cruzi DNA in cardiac lesions of Argentinean patients with end-stage chronic Chagas heart disease. Amer. J. trop. Med. Hyg., 70: 210-220, 2004.
- 27. SILBER, A.M.; BUA, J.; PORCEL, B.M.; SEGURA, E.L. & RUIZ, A.M. - Trypanosoma cruzi: specific detection of parasites by PCR in infected humans and vectors using a set of primers (BP1/BP2) targeted to a nuclear DNA sequence. Exp. Parasit., 85: 225-232, 1997.
- 28. SOUTO, R.P.; VARGAS, N. & ZINGALES, B. - Trypanosoma rangeli: discrimination from Trypanosoma cruzi based on a variable domain from the large subunit ribosomal RNA gene. Exp. Parasit., 91: 306-314, 1999.
- 29. STURM, N.R.; DEGRAVE, W.; MOREL, C. & SIMPSON, L. - Sensitive detection and schizodeme classification of Trypanosoma cruzi cells by amplification of kinetoplast minicircle DNA sequences: use in diagnosis of Chagas disease. Molec. Biochem. Parasit., 33: 205-214, 1989.
- 30. URREA, D.A.; CARRANZA, J.C.; CUBA, C.A. et al - Molecular characterisation of Trypanosoma rangeli strains isolated from Rhodnius ecuadoriensis in Peru, R. colombiensis in Colombia and R. pallescens in Panama, supports a co-evolutionary association between parasites and vectors. Infect. Genet. Evol., 5: 123-129, 2005.
- 31. VALLEJO, G.A.; GUHL, F.; CARRANZA, J.C. et al - kDNA markers define two major Trypanosoma rangeli lineages in Latin-America. Acta trop., 81: 77-82, 2002.
- 32. VALLEJO, G.A.; GUHL, F.; CARRANZA, J.C. et al - Parity between kinetoplast: DNA and miniexon gene sequences supports either clonal evolution or speciation in Trypanosoma rangeli strains isolated from R. colombiensis, R. pallescens and R. prolixus in Colombia. Infect. Genet. Evol., 3: 39-45, 2003.
- 33. VALLEJO, G.A.; GUHL, F.; CHIARI, E. & MACEDO, A.M. - Species specific detection of Trypanosoma cruzi and Trypanosoma rangeli in vector and mammalian hosts by Polymerase Chain Reaction amplification of kinetoplast minicircle DNA. Acta. trop., 72: 203-212, 1999.
- 34. VALLEJO, G.A.; MACEDO, A.M.; CHIARI, E. & PENA, S.D.J. - Kinetoplast DNA from Trypanosoma rangeli contains two distinct classes of minicircles with different size and molecular organization. Molec. Biochem. Parasit., 67: 245-253, 1994.
- 35. VALLEJO, G.A.; MARINKELLE, C.J.; GUHL, F. & DE SÁNCHEZ, N. - Comportamiento de la infección y diferenciación morfológica entre Trypanosoma cruzi y T. rangeli en el intestino del vector Rhodnius prolixus Rev. bras. Biol., 48: 577-587, 1988.
- 36. VARGAS, N.; SOUTO, R.P.; CARRANZA, J.C.; VALLEJO, G.A. & ZINGALES, B. - Amplification of a specific repetitive DNA sequence for Trypanosoma rangeli identification and its potential application in epidemiological investigations. Exp. Parasit., 96: 147-159, 2000.
- 37. VÁZQUEZ, M.; BEN-DOV, C.; LORENZI, H. et al - The short interspersed repetitive element of Trypanosoma cruzi, SIRE, is part of VIPER, an unusual retroelement related to long terminal repeat retrotransposons. Proc. nat. Acad. Sci. (Wash.), 97: 2128-2133, 2000.
- 38. VAZQUEZ, M.; LORENZI, H.; SCHIJMAN, A.G.; BEN-DOV, C. & LEVIN, M.J. - Analysis of the distribution of SIRE in the nuclear genome of Trypanosoma cruzi Gene, 239: 207-216, 1999.
- 39. VÁZQUEZ, M.P.; SCHIJMAN, A.G. & LEVIN, M.J. - A short interspersed repetitive element provides a new 3' acceptor site for trans-splicing in certain ribosomal P2 beta protein genes of Trypanosoma cruzi. Molec. Biochem. Parasit., 64: 327-336, 1994.
- 40. WINCKER, P.; BOSSENO, M.F.; BRITTO, C. et al - High correlation between Chagas' disease serology and PCR-based detection of Trypanosoma cruzi kinetoplast DNA in Bolivian children living in an endemic area. FEMS Microbiol. Lett., 124: 419- 423, 1994.
- 41. WINCKER, P.; BRITTO, C.; PEREIRA, J.B. et al - Use of a simplified polymerase chain reaction procedure to detect Trypanosoma cruzi in blood samples from chronic chagasic patients in a rural endemic area. Amer. J. trop. Med. Hyg., 51: 771-777, 1994.
-
42WORLD HEALTH ORGANIZATION - Control of Chagas disease Second report of the WHO Expert Committee. Wld Hlth Org. techn. Rep. Ser., (905) 39-40, 2002.
- 43. YEO, M.; ACOSTA, N.; LLEWELLYN, M. et al - Origins of Chagas disease: Didelphis species are natural hosts of Trypanosoma cruzi I and armadillos hosts of Trypanosoma cruzi II, including hybrids. Int. J. Parasit., 35: 225-233, 2005.
Correspondence to:
Publication Dates
-
Publication in this collection
19 Mar 2007 -
Date of issue
Feb 2007
History
-
Accepted
15 Aug 2006 -
Received
27 Apr 2006