Abstracts
Respiratory virus infections are the main cause of infant hospitalization and are potentially severe in children with congenital heart disease (CHD). Rapid and sensitive diagnosis is very important to early introduction of antiviral treatment and implementation of precautions to control transmission, reducing the risk of nosocomial infections. In the present study we compare different techniques in the diagnosis of respiratory viruses in CHD infants. Thirty-nine samples of nasopharyngeal aspirate were obtained from CHD infants with symptoms of respiratory infection. The Multiplex PCR (Seeplex® RV 12 ACE Detection) driven to the detection of 12 respiratory viruses was compared with the direct immunofluorescence assay (DFA) and PCR, both targeting seven respiratory viruses. The positivity found by DFA, Multiplex and PCR was 33.3%, 51.3% and 48.7%, respectively. Kappa index comparing DFA and Multiplex, DFA and PCR and PCR and Multiplex PCR was 0.542, 0.483 and 0.539, respectively. The concordance between techniques was considered moderate. Both Multiplex PCR (p = 0.001) and PCR (p = 0.002) detected significantly more respiratory virus than DFA. As the performance of the tests may vary, the combination of two or more techniques may increase diagnostic sensitivity favoring the diagnosis of co-infections, early introduction of antiviral therapy and implementation of appropriate measures.
Respiratory viruses; Congenital heart disease; Direct immunofluorescence assay; Multiplex PCR; PCR
Infecções respiratórias virais são a principal causa de hospitalização infantil e podem ser extremamente graves em crianças com cardiopatia congênita. O diagnóstico rápido e sensível é importante para a introdução precoce de tratamento antiviral e implantação de precauções para controle da transmissão, reduzindo o risco de infecções nosocomiais. Neste estudo, comparamos o desempenho de diferentes técnicas no diagnóstico de vírus respiratórios em crianças com cardiopatia congênita e sintomas respiratórios. Trinta e nove amostras de aspirado de nasofaringe foram obtidas de crianças com sintomas de infecção respiratória. Ensaio de PCR Multiplex que detecta 12 vírus respiratórios (Seeplex® RV 12 ACE Detection) foi comparado à Imunofluorescência Direta (IFD) e à PCR específica, ambas direcionadas a sete vírus. A positividade da IFD foi 33,3%, do Multiplex foi 51,3% e da PCR 48,7%. O índice kappa comparando IFD e Multiplex, IFD e PCR, e PCR e Multiplex foi, respectivamente, 0,542, 0,483 e 0,539, sendo a concordância considerada moderada. O Multiplex e a PCR detectaram significantemente mais vírus que a IFD (p < 0,0001 e 0,002, respectivamente). Como o desempenho dos testes varia o uso de mais de uma técnica pode aumentar a sensibilidade diagnóstica favorecendo a introdução precoce de terapia antiviral e implantação de medidas profiláticas
VIROLOGY
Identification of respiratory virus in infants with congenital heart disease by comparison of different methods
Identificação de vírus respiratórios em crianças com cardiopatia congênita por comparação de diferentes métodos
Tatiana Mitiko KanashiroI; Lucy Santos Vilas BoasI; Ana Maria ThomazII; Tania Regina Tozetto-MendozaI; Mônica SetsukoII; Clarisse Martins MachadoI
ILaboratório de Virologia, Instituto de Medicina Tropical de São Paulo, Universidade de São Paulo. Avenida Dr. Enéas de Carvalho Aguiar 470, 05403-000 São Paulo, SP, Brazil
IIINCOR - Unidade de Terapia Intensiva do Instituto do Coração, Hospital das Clínicas, Faculdade de Medicina, Universidade de São Paulo. Avenida Dr. Enéas de Carvalho Aguiar 44, 05403-900 São Paulo, SP, Brazil
Correspondence Correspondence to: Clarisse Martins Machado. Laboratório de Virologia, Instituto de Medicina Tropical de São Paulo, Universidade de São Paulo. Avenida Dr. Enéas de Carvalho Aguiar 470, 05403-000 São Paulo, SP, Brasil. Tel.: 55-11-3061-7020, Fax: 55-11-3063-2659. E-mail: clarimm@usp.br
SUMMARY
Respiratory virus infections are the main cause of infant hospitalization and are potentially severe in children with congenital heart disease (CHD). Rapid and sensitive diagnosis is very important to early introduction of antiviral treatment and implementation of precautions to control transmission, reducing the risk of nosocomial infections. In the present study we compare different techniques in the diagnosis of respiratory viruses in CHD infants. Thirty-nine samples of nasopharyngeal aspirate were obtained from CHD infants with symptoms of respiratory infection. The Multiplex PCR (Seeplex® RV 12 ACE Detection) driven to the detection of 12 respiratory viruses was compared with the direct immunofluorescence assay (DFA) and PCR, both targeting seven respiratory viruses. The positivity found by DFA, Multiplex and PCR was 33.3%, 51.3% and 48.7%, respectively. Kappa index comparing DFA and Multiplex, DFA and PCR and PCR and Multiplex PCR was 0.542, 0.483 and 0.539, respectively. The concordance between techniques was considered moderate. Both Multiplex PCR (p = 0.001) and PCR (p = 0.002) detected significantly more respiratory virus than DFA. As the performance of the tests may vary, the combination of two or more techniques may increase diagnostic sensitivity favoring the diagnosis of co-infections, early introduction of antiviral therapy and implementation of appropriate measures.
KEYWORDS: Respiratory viruses; Congenital heart disease; Direct immunofluorescence assay; Multiplex PCR; PCR.
RESUMO
Infecções respiratórias virais são a principal causa de hospitalização infantil e podem ser extremamente graves em crianças com cardiopatia congênita. O diagnóstico rápido e sensível é importante para a introdução precoce de tratamento antiviral e implantação de precauções para controle da transmissão, reduzindo o risco de infecções nosocomiais. Neste estudo, comparamos o desempenho de diferentes técnicas no diagnóstico de vírus respiratórios em crianças com cardiopatia congênita e sintomas respiratórios. Trinta e nove amostras de aspirado de nasofaringe foram obtidas de crianças com sintomas de infecção respiratória. Ensaio de PCR Multiplex que detecta 12 vírus respiratórios (Seeplex® RV 12 ACE Detection) foi comparado à Imunofluorescência Direta (IFD) e à PCR específica, ambas direcionadas a sete vírus. A positividade da IFD foi 33,3%, do Multiplex foi 51,3% e da PCR 48,7%. O índice kappa comparando IFD e Multiplex, IFD e PCR, e PCR e Multiplex foi, respectivamente, 0,542, 0,483 e 0,539, sendo a concordância considerada moderada. O Multiplex e a PCR detectaram significantemente mais vírus que a IFD (p < 0,0001 e 0,002, respectivamente). Como o desempenho dos testes varia o uso de mais de uma técnica pode aumentar a sensibilidade diagnóstica favorecendo a introdução precoce de terapia antiviral e implantação de medidas profiláticas.
doi: 10.1590/S0036-46652011000500001
INTRODUCTION
Acute respiratory tract infections (ARTIs) usually have a benign course but they are still the major determinant of death in developing countries and the most common cause of hospitalization of infants in developed countries. According to the World Health Organization, ARTIs were the leading cause of death in children under five years old in 200421. Previous studies in children younger than two years, with respiratory symptoms and clinical risk factors such as congenital heart disease (CHD), showed an augmented risk of severe infections, as demonstrated by greater morbidity and increased length of hospital stay13-15. Thus, the availability of a rapid and sensitive viral diagnostic assay is very important to establish appropriated therapeutic intervention, enabling physicians to make more accurate treatment decisions and reducing the use of unnecessary antibiotic therapy. Furthermore, rapid diagnosis favors the institution of specific precautions to minimize the risk of nosocomial infections16. In the last two decades, molecular techniques are improving, such as PCR (Polymerase Chain Reaction) or other amplification techniques of nucleic acid11. PCR and its variants are increasingly being used in routine laboratories because they are conceptually simple techniques, rapid, potentially very sensitive and specific and can be automated7,9,11. Several studies have evaluated the performance of in-house molecular techniques for diagnosis of respiratory viruses (RV)2,16,19. Commercial kits have also been evaluated and are considered rapid, sensitive and specific12. Currently, Seeplex® RV 12 ACE Detection, a commercial Multiplex PCR based on dual priming oligonucleotide (DPO) system for simultaneous detection of 12 respiratory viruses, has become available in clinical practice3,5,8,17,22. The aim of this study was to identify the respiratory viruses in CHD children with respiratory symptoms and to compare the performance of different techniques.
MATERIAL AND METHODS
1. ETHICAL CONSIDERATIONS: The current study is part of an ongoing project named "Respiratory Syncytial Virus (RSV) and Congenital Heart Disease", conducted at InCor - HCFMUSP (Hospital das Clínicas, Faculdade de Medicina, Universidade de São Paulo). The main project was designed to evaluate the prevalence of RSV in CHD children younger than four years, receiving or not RSV prophylaxis with palivizumab. The project was approved by the Ethics Committee of InCor - HCFMUSP. Respiratory samples were obtained after signature of the consent form by parents or guardians.
2. POPULATION AND SAMPLE SELECTION: We selected 39 samples of nasopharyngeal aspirate (NPA) obtained from 35 CHD infants (four children had two samples) with symptoms of upper and/or lower respiratory tract infections, assisted at the Emergency Room and Pediatric Intensive Care Unit of InCor - HCFMUSP. Thirteen samples had tested positive by direct immunofluorescence assay (DFA), being five (12.8%) positive for RSV, three (7.7%) for influenza A (INF A), two (5.1%) positive for adenovirus (ADV), two (5.1%) for parainfluenza 3 (PIV3) and one (2.6%) for parainfluenza 1(PIV1). The remaining 26 samples had tested negative by DFA.
3. RESPIRATORY SAMPLES: Five milliliters of saline solution 0.9% were instilled into each nostril and aspirated in a sterile collector. The samples, kept at 4 °C, were promptly sent to the laboratory and processed for DFA within two hours. An aliquot was stored at -70 ºC for subsequent PCRs (Multiplex PCR, in-house PCR and Real Time PCR).
4. DFA: The DFA using specific monoclonal antibodies (Kit Light DiagnosticsTM Respiratory DFA Viral Screening & Identification - Millipore, Bilerica, MA, USA) was performed for detection of influenza A and B, parainfluenza 1, 2 and 3, adenovirus and RSV in accordance with the manufacturer´s instructions.
5. RNA EXTRACTION AND REVERSE TRANSCRIPTION: RNA extraction was performed with the QIAamp®Viral RNA Mini Kit 250 (QIAGEN, Hilden, Germany) and the cDNA synthesis was performed using kit High-Capacity cDNA Reverse Transcription (Applied Biosystems, Foster City, CA, USA) in accordance with the manufacturer´s protocol. The products were stored at -70 °C before molecular testing. The same extraction method was used for adenovirus detection, and the ADV transcripts were identified.
6. MULTIPLEX PCR: The kit Seeplex® RV 12 ACE Detection (Seegene, Seoul, Korea), was used to detect 12 respiratory virus: ADV, human metapneumovirus (hMPV), coronavirus (hCoV) 229E/NL63, hCoV OC43/HKU1, PIV 1, 2 and 3, INF A and INF B, RSV A and B and rhinovirus (HRV) A/B, according to the manufacturer´s instructions with some modifications. GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA) was the thermal cycler used. Positive and negative controls were included in each reaction. PCR was performed with a final reaction volume of 20.0 µL containing 3.0 µL of cDNA. The amplified PCR products were separated on 2% agarose gels (Invitrogen, Carlsbad, CA) stained with ethidium bromide (BioAgency, São Paulo, BR). Three microliters of the amplified product and negative control; 1.0 µL positive control and 5.0 µL reference marker and ladder 100 bp (Fermentas, Burlington, CA) were applied into the gel. Specific fragment size was revealed by UV transillumination and recorded by AlphaImager® EC System, (Alpha Innotech Corporation, San Leandro, CA). The type of respiratory virus was identified by comparison with the reference specific fragment size provided by the manufacturer.
7. PCR SPECIFIC FOR EACH VIRUS (PCR):
7.1.IN-HOUSE PCR: This technique was performed to detect adenovirus, human metapneumovirus and RSV A and B. References and primers are described in Table 1 with few modifications1,6,19. Known positive samples and DEPC water were used as positive and negative controls, respectively, in each reaction. Mastercycler Gradient Eppendorf (Eppendorf, Hamburg, Germany) was the thermal cycler used. The amplified PCR products were separated on 1.5% agarose gel (Invitrogen, Carlsbad, CA) stained with ethidium bromide (BioAgency, São Paulo, BR). For amplification reaction for adenovirus and human metapneumovirus, they were applied on gel 10.0 µL of amplified product and 5.0 µL of ladder 100 bp (Fermentas, Burlington, CA). For RSV reaction they were applied with 10.0 µL of PCR product with 2.0 µL of bromophenol blue. Specific fragment size was revealed as described above.
7.1.1. Adenovirus: NestedPCR was performed. For first-round, the PCR mixture (50.0 µL) contained 3.0 µL of cDNA, 1 x reaction buffer (50 mM KCl, 10 mM Tris-HCl), 0.2 mM deoxynucleotide (dGTP, dATP, dTTP and dCTP), 1.5 mM MgCl2, 0.2 mM of each primer (hex1deg and hex2deg) (Table 1), Taq polimerase 2 U/reaction (Invitrogen, Carlsbad, CA), glycerol 4.5% and Cresol Red 0.25 µg/µL. PCR cycle conditions consisted of three minutes at 94 °C and 35 cycles of 30 seconds at 94 °C, 30 seconds at 55 °C, one minute at 72 °C and a final step for five minutes at 72 °C. For the second-round PCR, 3.0 µL of the firstround PCR product was used and the same conditions were applied, but another set of primers was used: nehex3deg (5'-GCCCGYGCMACIGAIACSTACTTC- 3') and nehex4deg (5'-CCYACRGCCAGIGTRWAICGMRCYTTGTA-3') (Table 1).
7.1.2. Human Metapneumovirus: PCR was conducted in a final reaction volume of 25.0 µL containing 5.0 µL of cDNA, 1x buffer (50 mM KCl, 10 mM Tris-HCl), 0.2 mM deoxynucleotide (dGTP, dATP, dTTP and dCTP), 1.5 mM MgCl2, 0.1 µM of each primer, glycerol 4.5%, Cresol Red 0.25µg/µL and Taq polymerase 1 U/reaction (Invitrogen, Carlsbad, CA). The PCR protocol was 95 ºC for two minutes; 40 cycles at 94 ºC for one minute, 55 ºC for one minute and 72 ºC for one minute, followed by 10 minutes at 72 ºC.
7.1.3. RSV: PCR amplification was performed in a final reaction volume of 25.0 µL using 5.0 µL of cDNA, 1x buffer (50 mM KCl, 10 mM Tris-HCl), 0.2 mM deoxynucleotide (dGTP, dATP, dTTP and dCTP), 1.5 mM MgCl2, 2.0 µM of each primer and Taq polimerase 2.5 U/reaction (Invitrogen, Carlsbad, CA). An initial step of 94 ºC for two minutes was performed, followed by a total of 35 cycles at 94 ºC for one minute, 65 ºC for one minute and 72 ºC for one minute, with final cycle at 72 ºC for seven minutes.
7.2. REAL TIME PCR: This technique was performed to detect influenza A and B and parainfluenza 3. References, primers and probes used are listed in Table 2, with few modifications18,20. Known positive samples and DEPC water were used as positive and negative controls, respectively, in each reaction. Amplification and detection was realized using ABI 7300 Real Time PCR System (Applied Biosystem, Foster City, California, USA).
7.2.1. Influenza: For INF A, one reaction (25.0 µL) was carried out with 5.0 µL of cDNA, 1x TaqMan® Universal PCR Master Mix (Applied Biosystems, Foster City, California), 0.9 µM of primer INFA-1, 0.45 µM of each primer INFA-2 and 3 and 0.1 µM of probe. Thermal conditions and the protocol for INF B were conducted according to VAN ELDEN et al., 200120.
7.2.2. Parainfluenza 3: PCR was conducted in a final reaction volume of 25.0 µL with 3.0 µL of cDNA, 1x TaqMan® Universal PCR Master Mix (Applied Biosystems, Foster City, California), 0.3 µM of each primer and 0.2 µM of probe. PCR cycle conditions consisted of two minutes at 50 °C and followed by 10 minutes at 95 °C and 45 cycles of 15 seconds at 95 ºC and one minute at 60 ºC. PIV 3 primers were described by SOUZA (Experimental Research Center, IIEP, Albert Einstein Hospital, unpublished data, personal communication)18.
8. STATISTICAL ANALYSIS: Positive results for hCoV 229E/NL63, hCoV OC43/HKU1 and HRV A/B identified by Multiplex PCR (Seeplex) were not considered in the comparison between techniques. Kappa index was used to compare DFA, Multiplex PCR and PCR. PCR was considered the gold standard for comparison between techniques. Statistical analysis was performed using SPSS software,
version 15.0.
RESULTS
A total of 39 samples of nasopharyngeal aspirate were taken from 35 infants with CHD and symptoms of respiratory infection. Of these, 25 (64.1%) tested positive by at least one of the techniques. Thirteen samples (33.3%) were positive for DFA, 20 (51.3%) were positive for Multiplex PCR and 19 (48.7%) for PCR (Table 3).
As shown in Table 3, DFA and Multiplex PCR showed greater concordance, with 30 (77%) concordant results, being 18 (46.2%) negative and 12 (30.8%) positive (Kappa = 0.542). The greatest concordance between these techniques occurred in the diagnosis of RSV and the smallest concordance in the diagnosis of ADV. PCR and DFA showed 29 (74.4%) concordant results, with 18 (46.2%) negative and 11 (28.2%) positive (Kappa = 0.483). Equally, RSV diagnosis had the greatest concordance and ADV the smallest concordance. Finally, PCR and Multiplex PCR showed 15 (38.5%) negative and positive concordant results (Kappa = 0.539). The greatest concordance was observed in the diagnosis of ADV and smallest in the diagnosis of RSV.
Considering PCR as gold standard, the sensitivity and specificity of DFA were 57.9% and 90.0%, respectively, with kappa index 0.483 (p = 0.002). Regarding Multiplex PCR, the sensitivity and specificity were 78.9% and 75.0%, respectively, with kappa index 0.539 (p = 0.001).
The RVs more frequently identified were RSV and adenovirus, considering all techniques. Table 4 shows the positivity of each technique according to the respiratory virus analyzed.
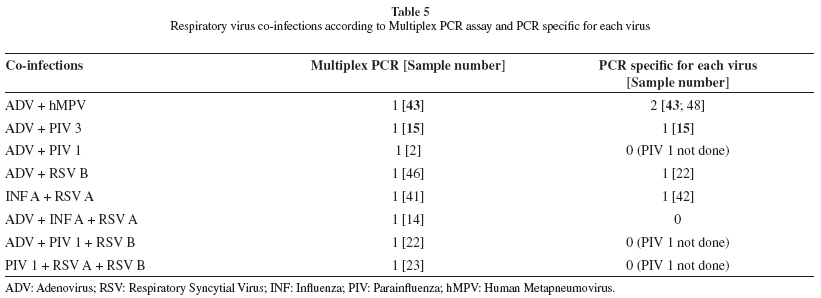
Multiplex PCR and PCR detected eight and five co-infections, respectively (Table 5). Only two showed concordance in both techniques: ADV and human metapneumovirus, and ADV and parainfluenza 3. No co-infection was detected by DFA.
DISCUSSION
Viral respiratory tract infections may be potentially severe, especially in children with congenital heart disease13-15; so it is essential a sensitive and rapid test to detect pathogens.
In this study, the use of three different techniques favored the detection of at least one respiratory virus in approximately 64% of samples.
In the diagnosis of respiratory viruses, DFA has been more frequently used in several clinical laboratories, as the technique can provide specific diagnosis in a few hours, so several commercial kits should be on hand. However, the monoclonal antibodies available are directed to a limited number of viruses, excluding the ones most recently discovered. Moreover, this technique has a lower sensitivity when compared with molecular techniques11. Also, the reader subjectivity, the need for specimens with the appropriate number of cells, and the impossibility of automation are other problems associated with the DFAs.
Recently, a new molecular technique has been used, based on a dual priming oligonucleotide (DPO) system, developed by Chun et al.4. The system uses a primer that contains two regions joined by a polydeoxyinosine link that allow a wide range of annealing temperatures, providing high specificity, and preventing false positive results.
As observed in other comparative studies, Multiplex PCR based on DPO system has been considered a sensitive8,17,22 and specific technique8 for detection of multiple respiratory viruses and consequently, an effective tool for use in a clinical laboratory5, favoring early diagnosis.
In the present study, both Multiplex and PCR specific detected significantly more respiratory viruses in comparison to DFA (p < 0.0001 and p = 0.002, respectively).
Other studies have shown similar results. Multiplex PCR and DFA positivity was 51.3% and 33.3%, respectively, similar to those found by Yoo et al. (56% and 33%)22. However, the 48.7% positivity of our PCR techniques (in-house and Real Time) was lower than the 69% positivity obtained by Bruijnesteijn van Coppenraet et al.3 who used Real Time PCR.
According to kappa index, concordance between tests was considered moderate. Among the 13 positive samples by DFA only one (influenza A) did not agree with any molecular technique. All other positive results were concordant at least with Multiplex PCR. We cannot be sure about RSV type concordance because DFA does not discriminate between RSV A and RSV B.
Among the 26 negative samples by DFA, seven (26.9%) and six (23.1%) samples were positive by Multiplex PCR and PCR, respectively. These results were expected since molecular techniques have a higher capability of detection even in clinical samples with lower titer of virus8,10. Also, several co-infections were detected using molecular techniques but not by DFA. Multiplex PCR detected eight co-infections, accounting for 40% of positive samples. Other studies have found between 9 to 18% of co-infections with the same technique3,8,17,22. PCR diagnosed five (26.3% of positive samples) co-infections. Probably, DFA has not been detected due to low sensitivity of the technique, especially in relation to adenovirus, although some authors attribute this to the subjective IF readouts of this technique11. The ability to detect co-infections is extremely important for providing data for studies of clinical importance of co-infections, to investigate whether certain individual groups are at greater risk for co-infections or whether they result in a poorer outcome for the patient. In addition, multiplex tests can contribute to understanding the epidemiology of viral infections and provide new information on seasonality, geographical distribution and risk groups11.
In the present study, the concordance between tests was considered moderate and molecular techniques detected significantly more virus than DFA. Multiplex PCR is a faster and less laborious technique than PCR specific for each virus, because it is able to detect 12 respiratory viruses in only two amplification reactions. In contrast, PCR would require one reaction for each virus. On the other hand, Multiplex PCR requires greater care in gel load and electrophoresis for a discriminating interpretation of the results.
In conclusion, the use of more than one technique can increase the sensitivity in respiratory virus detection. The Seeplex® RV 12 ACE Detection kit showed a good performance and may contribute to early diagnosis of RV infections, especially in the presence of co-infections and in patients at higher risk of complications. Other studies are necessary to demonstrate the cost-effectiveness of this assay in comparison with other rapid techniques.
ACKNOWLEDGEMENTS
The study was partially supported by Abbott. Kit Seeplex was kindly provided by Bioeasy Diagnóstica.
Received: 1 April 2011
Accepted: 22 August 2011
- 1. Allard A, Albinsson B, Wadell G. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. J Clin Microbiol. 2001;39:498-505.
- 2. Bharaj P, Sullender WM, Kabra SK, Mani K, Cherian J, Tyagi V, et al. Respiratory viral infections detected by multiplex PCR among pediatric patients with lower respiratory tract infections seen at an urban hospital in Delhi from 2005 to 2007. Virol J. 2009;6:89.
- 3. Bruijnesteijn van Coppenraet LE, Swanink CM, van Zwet AA, Nijhuis RH, Schirm J, Wallinga JA, et al. Comparison of two commercial molecular assays for simultaneous detection of respiratory viruses in clinical samples using two automatic electrophoresis detection systems. J Virol Methods. 2010;169:188-92.
- 4. Chun JY, Kim KJ, Hwang IT, Kim YJ, Lee DH, Lee IK, et al. Dual priming oligonucleotide system for the multiplex detection of respiratory viruses and SNP genotyping of CYP2C19 gene. Nucleic Acids Res. 2007; 35:e40.
- 5. Drews SJ, Blair J, Lombos E, DeLima C, Burton L, Mazzulli T, et al. Use of the Seeplex RV detection kit for surveillance of respiratory viral outbreaks in Toronto, Ontario, Canada. Ann Clin Lab Sci. 2008;38:376-9.
- 6. Falsey AR, Erdman D, Anderson LJ, Walsh EE. Human metapneumovirus infections in young and elderly adults. J Infect Dis. 2003;187:785-90.
- 7. Harrison TJ. The polymerase chain reaction--a time of transition from research to routine. J Clin Pathol. 1998;51:491-2.
- 8. Kim SR, Ki CS, Lee NY. Rapid detection and identification of 12 respiratory viruses using a dual priming oligonucleotide system-based multiplex PCR assay. J Virol Methods. 2009;156:111-6.
- 9. Klapper PE, Jungkind DL, Fenner T, Antinozzi R, Schirm J, Blanckmeister C. Multicenter international work flow study of an automated polymerase chain reaction instrument. Clin Chem. 1998;44:1737-9.
- 10. Liolios L, Jenney A, Spelman D, Kotsimbos T, Catton M, Wesselingh S. Comparison of a multiplex reverse transcription-PCR-enzyme hybridization assay with conventional viral culture and immunofluorescence techniques for the detection of seven viral respiratory pathogens. J Clin Microbiol. 2001;39:2779-83.
- 11. Mahony JB. Detection of respiratory viruses by molecular methods. Clin Microbiol Rev. 2008; 21:716-47.
- 12. Marshall DJ, Reisdorf E, Harms G, Beaty E, Moser MJ, Lee WM, et al. Evaluation of a multiplexed PCR assay for detection of respiratory viral pathogens in a public health laboratory setting. J Clin Microbiol. 2007;45:3875-82.
- 13. Medrano C, Garcia-Guereta L, Grueso J, Insa B, Ballesteros F, Casaldaliga J, et al. Respiratory infection in congenital cardiac disease. Hospitalizations in young children in Spain during 2004 and 2005: the CIVIC Epidemiologic Study. Cardiol Young. 2007;17:360-71.
- 14. Medrano López C, García-Guereta Silva L. Infecciocnes respiratorias y cardiopatías: dos estaciones del studio CIVIC. An Pediatr (Barc). 2007;67:329-36.
- 15. Medrano López C, García-Guereta Silva L, Lirio Casero J, García Pérez J, Grupo CIVIC Grupo de Trabajo de Infecciones de la Sociedad Espańola de Cardiología Pediátrica y Cardiopatías Congénitas. Infecciones respiratorias, syndrome de Down y cardiopatías congenitas: estudio CIVIC 21. An Pediatr (Barc). 2009;71:38-46.
- 16. Reis AD, Fink MC, Machado CM, Paz JPJr, Oliveira RR, Tateno AF, et al. Comparison of direct immunofluorescence, conventional cell culture and polymerase chain reaction techniques for detecting respiratory syncytial virus in nasopharyngeal aspirates from infants. Rev Inst Med Trop Sao Paulo. 2008;50:37-40.
- 17. Roh KH, Kim J, Nam MH, Yoon S, Lee CK, Lee K, et al. Comparison of the Seeplex reverse transcription PCR assay with the R-mix viral culture and immunofluorescence techniques for detection of eight respiratory viruses. Ann Clin Lab Sci. 2008;38:41-6.
- 18. Souza AV. Experimental Research Center, IIEP, Albert Einstein, unpublished data, personal communication. Săo Paulo.
- 19. Sullender WM, Sun LR, Anderson LJ. Analysis of respiratory syncytial virus genetic-variability with amplified cDNAs. J Clin Microbiol. 1993;31:1224-31.
- 20. van Elden LJR, Nijhuis M, Schipper P, Schuurman R, van Loon AM. Simultaneous detection of influenza viruses A and B using real-time quantitative PCR. J Clin Microbiol. 2001;39:196-200.
-
21WHO. The global burden of disease. 2004 update. Geneva: World Health Organization; 2008.
- 22. Yoo SJ, Kuak EY, Shin BM. Detection of 12 respiratory viruses with two-set multiplex reverse transcriptase-PCR assay using a dual priming oligonucleotide system. Korean J Lab Med. 2007;27:420-7.
Publication Dates
-
Publication in this collection
13 Oct 2011 -
Date of issue
Oct 2011
History
-
Received
01 Apr 2011 -
Accepted
22 Aug 2011