Abstracts
The objective of this study is to identify subtypes of Human Immunodeficiency Virus type 1 (HIV-1) and to analyze the presence of mutations associated to antiretroviral resistance in the protease (PR) and reverse transcriptase (RT) regions from 48 HIV-1 positive treatment naïve patients from an outpatient clinic in Maringá, Paraná, Brazil. Sequencing was conducted using PR, partial RT and group-specific antigen gene (gag) nested PCR products from retrotranscribed RNA. Transmitted resistance was determined according to the Surveillance Drug Resistance Mutation List (SDRM) algorithm. Phylogenetic and SimPlot analysis of concatenated genetic segments classified sequences as subtype B 19/48 (39.6%), subtype C 12/48 (25%), subtype F 4/48 (8.3%), with 13/48 (27.1%) recombinant forms. Most recombinant forms were B mosaics (B/F 12.5%, B/C 10.4%), with one C/F (2.1%) and one complex B/C/F mosaic (2.1%). Low levels of transmitted resistance were found in this study, 2/48 (2.1% to NRTIs and 2.1% for PI). This preliminary data may subsidize the monitoring of the HIV evolution in the region.
Primary resistance; Transmitted resistance; HIV-1; Naïve patients; Subtypes; Brazil
O objetivo foi identificar subtipos do Vírus da Imunodeficiência Humana tipo-1 (HIV-1) e analisar a presença de mutações/polimorfismos nas regiões da protease (PR) e transcriptase reversa (TR) de 48 pacientes virgens de tratamento atendidos no município de Maringá, Paraná, Brasil. O sequenciamento foi conduzido usando produtos de nested PCR dos genes da PR, TR parcial e group-specific antigen gene (gag) de RNA retrotranscrito. A interpretação da resistência transmitida foi realizada segundo o algoritmo Surveillance Drug Resistance Mutation List (SDRM). As análises filogenética e SimPlot dos segmentos concatenados classificaram as sequências como subtipo B 19/48 (39,6%), subtipo C 12/48 (25%), subtipo F 4/48 (8,3%), com 13/48 (27,1%) formas recombinantes. A maioria das formas recombinantes era mosaicos B (B/F 12,5%, B/C 10,4%), com um C/F (2,1%) e um mosaico complexo B/C/F (2,1%). A prevalência de resistência transmitida foi de 4,2% (2,1% para ITRN e 2,1% para IP). Baixos níveis de resistência transmitida foram encontrados nesse estudo, 2/48 (2,1% para INTR e 2,1% para IP). Esses achados, embora preliminares, podem contribuir no monitoramento da epidemia de HIV na região.
HIV
Genetic diversity and primary resistance among HIV-1-positive patients from Maringá, Paraná, Brazil
Diversidade genética e resistência primária entre pacientes HIV-1-positivos de Maringá, Paraná, Brasil
Karine Vieira GasparetoI; Flávia Myrian Martins de Almeida MelloI; José Ricardo Colleti DiasII; Vera Alice Fernandes MeneguettiII; Marta Evelyn Giansante StortiII; João Leandro de Paula FerreiraIII; André Minhoto LançaIII; Rosângela RodriguesIII; Luis Fernando de Macedo BrígidoIII; Jorge Juarez Vieira TeixeiraIV; Dennis Armando BertoliniIV
IMestrado em Ciências da Saúde, Universidade Estadual de Maringá, Maringá, PR, Brazil
IIServiço de Atendimento Especializado em DST/aids (SAE) de Maringá, Secretaria de Saúde de Maringá, Maringá, PR, Brazil
IIILaboratório de Retrovírus, Centro de Virologia, Instituto Adolfo Lutz, São Paulo, SP, Brazil
IVDepartamento de Análises Clínicas e Biomedicina, Universidade Estadual de Maringá, Maringá, PR, Brazil
Correspondence to Correspondence to: Dennis Armando Bertolini Departamento de Análises Clínicas e Biomedicina Universidade Estadual de Maringá Avenida Colombo 5790, 87020-900 Maringá, Paraná, Brasil Tel. + 00 55 44 3011 4878; Fax: + 00 55 44 3011 4860 E-mail: dabertolini@uem.br
SUMMARY
The objective of this study is to identify subtypes of Human Immunodeficiency Virus type 1 (HIV-1) and to analyze the presence of mutations associated to antiretroviral resistance in the protease (PR) and reverse transcriptase (RT) regions from 48 HIV-1 positive treatment naïve patients from an outpatient clinic in Maringá, Paraná, Brazil. Sequencing was conducted using PR, partial RT and group-specific antigen gene (gag) nested PCR products from retrotranscribed RNA. Transmitted resistance was determined according to the Surveillance Drug Resistance Mutation List (SDRM) algorithm. Phylogenetic and SimPlot analysis of concatenated genetic segments classified sequences as subtype B 19/48 (39.6%), subtype C 12/48 (25%), subtype F 4/48 (8.3%), with 13/48 (27.1%) recombinant forms. Most recombinant forms were B mosaics (B/F 12.5%, B/C 10.4%), with one C/F (2.1%) and one complex B/C/F mosaic (2.1%). Low levels of transmitted resistance were found in this study, 2/48 (2.1% to NRTIs and 2.1% for PI). This preliminary data may subsidize the monitoring of the HIV evolution in the region.
Keywords: Primary resistance; Transmitted resistance; HIV-1; Naïve patients; Subtypes; Brazil.
RESUMO
O objetivo foi identificar subtipos do Vírus da Imunodeficiência Humana tipo-1 (HIV-1) e analisar a presença de mutações/polimorfismos nas regiões da protease (PR) e transcriptase reversa (TR) de 48 pacientes virgens de tratamento atendidos no município de Maringá, Paraná, Brasil. O sequenciamento foi conduzido usando produtos de nested PCR dos genes da PR, TR parcial e group-specific antigen gene (gag) de RNA retrotranscrito. A interpretação da resistência transmitida foi realizada segundo o algoritmo Surveillance Drug Resistance Mutation List (SDRM). As análises filogenética e SimPlot dos segmentos concatenados classificaram as sequências como subtipo B 19/48 (39,6%), subtipo C 12/48 (25%), subtipo F 4/48 (8,3%), com 13/48 (27,1%) formas recombinantes. A maioria das formas recombinantes era mosaicos B (B/F 12,5%, B/C 10,4%), com um C/F (2,1%) e um mosaico complexo B/C/F (2,1%). A prevalência de resistência transmitida foi de 4,2% (2,1% para ITRN e 2,1% para IP). Baixos níveis de resistência transmitida foram encontrados nesse estudo, 2/48 (2,1% para INTR e 2,1% para IP). Esses achados, embora preliminares, podem contribuir no monitoramento da epidemia de HIV na região.
INTRODUCTION
In Brazil, 492,581 cases of AIDS were identified between 1980 and 2010. The prevalence of HIV in the population is estimated to be around 0.6%. The distribution of the cases according to regions, in 2009, showed that 38.2% of the cases are in the southeastern region, followed by the northeast (21.9%), south (21.1%), north (11.1%) and center-west (7.7%). Although more cases were recorded in the southeastern region, the south has a higher prevalence of AIDS cases, 12.6/100,000 inhabitants. The city of Maringá, situated in the north-central part of the State of Paraná, had 34.9 reported cases of AIDS per 100,000 inhabitants in 20093. In 2010, a total of 201,279 patients were receiving antiretroviral therapy in Brazil, 45,308 in the south region and 10,821 in Paraná4.
In newly infected individuals, treatment may be compromised because of the possibility of transmission of viral variants that are resistant to antiretroviral drugs, from some treated patients who had been exposed to antiretrovirals44. Several studies involving antiretroviral treatment-naïve patients have been conducted independently in different Brazilian cities, and the reported rates of primary resistance range from 1.4 to 8.3%7,10,20. The first large-scale evaluation of HIV diversity and transmitted resistance that included samples from southern Brazil, including Rio Grande do Sul (n = 139) and Paraná (n = 147), showed prevalence rates of 44.88% and 29.79% for subtype C, 29.92% and 63.83% for B, 3.15% and 4.26% for F, and 22.05% and 21.3% for recombinant forms, respectively8.
The present study analyzed sequences of the protease, reverse transcriptase and gag genes for the evaluation of HIV-1 viral subtypes and the identification of mutations/polymorphisms related to antiretroviral resistance among treatment-naïve patients seen at the STD/AIDS Specialty Clinic in the city of Maringá, Paraná, Brazil.
MATERIALS AND METHODS
Study population: HIV-1-positive treatment-naïve patients (n = 61) seen at the STD/AIDS Specialty Clinic in Maringá, from April through June 2009 were invited to participate in this study. The study was approved by the Ethics Committee on research involving humans of the Universidade Estadual de Maringá (CAAE No. 0047.0.093.000-09; protocol 126/2009), and written informed consent was obtained from each patient. Sociodemographic data were collected from the patient records.
Calculation of sample size: The sample size was defined in accordance with the protocol of the World Health Organization (WHO) HIV Drug-Resistance Threshold Survey (HIV DR THS)27.
CD4+ and CD8+ T cell counts and quantification of HIV-1 RNA: The number of CD4+ and CD8+ T cells was determined by flow cytometry (BD TrucountTM Tubes) in a FACSCalibur flow cytometer (Becton-Dickinson, New Jersey, USA). Viral load was evaluated by the branched DNA assay (VERSANT® HIV-1 RNA 3.0 Assay) in a 340 bDNA Analyzer (Bayer HealthCare, New York, USA) according to the manufacturer's instructions. The quantification limit of the assay ranges from 50 to 500,000 copies/mL plasma.
RNA extraction and synthesis of cDNA: HIV-1 RNA was extracted using the QIAmp® Viral RNA Mini Handbook extraction kit (QIAGEN, Courtaboeuf, France). cDNA was retrotranscribed using superscript II (Invitrogen, USA).
Amplification of the PR/RT region: In the first step of the nested PCR, a partial Pol gene was amplified using the K1 and K2 primers (PR/RT, 1,200 bp)22. The second reaction allowed us to amplify regions of the complete PR and RT genes, separately. The DP10 and DP11 primers (400 bp)31 were used to amplify the protease region, and the F1 and F2 primers (800 bp)16 to amplify the RT region.
Amplification of the gag gene: In the first step of the nested PCR, the H1G777 and H1P202 primers12 were used to amplify a region of the gag gene (1,100 bp). The second step of the nested PCR amplified a fragment encoding p24 (460 bp). The H1gag1584 and g17 primers12 were used for this amplification.
Sequencing: The PCR products were sequenced as described previously33 using the following primers: G17 and H1gag1584 for the gag gene, DP10 and DP11 for the protease gene, and F1 and F2 for the RT gene. Sequencing was carried out using fluorescent-labeled dideoxynucleotides (ddNTPs) and the ABI Prism BigDyeTM Terminator Ready Reaction Cycle Sequencing kit, version 3.1 (Applied Biosystems, Foster City, CA, USA). After precipitation of the reaction product, the samples were denatured and sequenced in an automated ABI 3100 sequencer (Applied Biosystems).
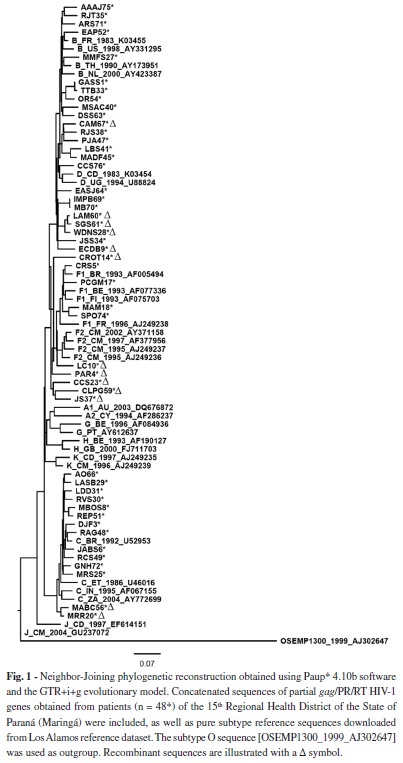
Sequence analysis: The sequences obtained were first analyzed using the Phred-Phrap program (http://asparagin.cenargen.embrapa.br/phph/) for assessment of data quality and construction of the consensus sequences using bases with a score > 2039. The sequences were aligned with the Muscle program, version 3.614 and edited using the Bioedit Sequence Alignment Editor, version 7.0.9.019. The phylogenetic signal was obtained with the DAMBE program, version 5.0.3242, and genetic diversity and distance were calculated with the MEGA 4 program (Molecular Evolutionary Genetics Analysis)38. The Tamura-Nei model was used for signal and diversity calculations, while the Kimura two-parameter model was employed in the distance analysis. To increase the gag phylogenetic signal, PR and RT sequences from each patient were concatenated using MS Excel and BioEdit software. Subtypes were screened with the REGA HIV-1 Subtyping tool (http://www.bioafrica.net/rega-genotype/html/subtypinghiv.html)28 and NCBI genotyping (http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi)32, while phylogenetic reconstruction, performed with PAUP* 4.10b software37, and bootscanning (SimPlot version 3.5.125) employing a 200-bp window and a 20-bp stepsize, using 500 replicates, were used to confirm subtypes and unique recombinant forms (URF), respectively. Neighbor-Joining (NJ) and Maximum Likelihood (ML) trees were obtained using the GTR+i+g model as determined using ModelTest3.7. Bootstrap values were obtained using 1000 replicates to test the reliability of the tree. The reference sequences were obtained from the Los Alamos National database.
Algorithms used for the analysis of resistance-associated mutations and polymorphisms: To classify and identify polymorphisms and mutations associated with resistance to ARVs, the FASTA sequences of the PR and RT were analyzed using the freely available SDRM-2009 algorithm available in the Calibrated Population Resistance tool (CPR), version 6.0 beta (http://cpr.stanford.edu/cpr/servlet/CPR)17. The SDRM algorithm was applied to determine the prevalence of primary ARV resistance mutations, using a list of drug-resistance mutations that provide an estimate of resistance transmission according to the WHO guidelines.
Statistical analysis: Data were analyzed using the STATISTICA 7.135 and Open Source Epidemiologic Statistics for Public Health (OpenEpiversion 2.311) programs. The χ2 test and Fisher's exact test were used to compare categorical variables, and ANOVA was applied to compare continuous variables. A p value < 0.05 was considered to be statistically significant.
RESULTS
Samples from 61 patients were submitted to PR, RT and gag PCR. Of these, the PR and RT regions were amplified in 48 (78.7%) and the gag gene in 46 (75.4%), and these isolates were sequenced. Sociodemographic and baseline characteristics of the patients are summarized in Table 1.
Sequences analysis of the 48 isolates for subtype classification showed that subtypes B (n = 19, 39.6%) and C (n = 12, 25%) were the most prevalent, followed by subtype F (n = 4, 8.3%) (Fig. 1). The frequency of recombinant forms was 27.1% (n = 13), including subtypes B/F in six (12.5%), B/C in five (10.4%) and F/C in one (2.1%). One complex genome, possibly including genomes B/C/F, was also observed (2.1%).
The gag region was not amplified in two isolates classified as subtype B according to both the PR and RT regions, and considered here as B (Table 2). The HIV-1 genetic distance was 9.2 ± 0.7% for RT, 9.5 ± 1.0% for PR and 10.2 ± 1.1% for gag.
Table 3 shows the epidemiological data for the 48 treatment-naïve patients according to viral subtype. Statistical analysis revealed a significant difference in viral load, with higher values being observed in patients harboring recombinant forms of the virus (p < 0.01). Although the difference was significant, a broader evaluation is necessary to confirm the biological significance of our findings. Quantification of CD4+ T cells showed no statistically significant difference (p = 0.773).
According to the SDRM algorithm, the prevalence of primary resistance mutations was 4.2%, with 2.1% to PIs (V82L and L90M) and 2.1% to NRTIs (K219N). Both mutations detected in the protease region belonged to a single patient.
The protease polymorphisms most frequently found in the 48 isolates sequenced were M36I (62.5%), I15V (60.4%), L63P (43.7%), E35D (35.4%) and L10I/V (8.3%). The most frequent RT polymorphisms were R211K (58.3%), V35T (45.8%), K122E (41.6%), Q207E (35.4%), I135T (33.3%) and V118I (6.2%).
DISCUSSION
This study describes HIV-1 molecular epidemiology findings from Maringá, Paraná. Although many studies have treated this subject, few were conducted outside major cities, and this is the first to describe the situation in this region. We assessed 61 antiretroviral-naïve patients from the local AIDS clinic, and succeeded in obtaining molecular data from 48 (79%). The main factor associated with failure of PCR amplification was a low plasma viral RNA load. With a mean plasma viral load of 4.2 (± 0.7) log10/mL among the 48 patients who were successfully evaluated, patients with sequences that were not amplified in this study showed a mean number of viral RNA copies (log10) of 3.0 (± 1.6)/mL plasma (< 50 to 171,563 RNA copies/mL plasma). Ten of the 13 non-amplified isolates contained < 5000 RNA copies/mL plasma.
Subtype C was detected in 25%; other studies analyzing samples from patients of the southern region have observed similarly high prevalences of subtype C7,8,24,34. For instance, a study conducted in Santa Catarina (n = 80) reported a high prevalence of subtype C (48.75%), which increased to 71.25% when the recombinant viruses containing subtype C-related sequences were included in the analysis24. In a study with samples from a voluntary testing site in Curitiba, Paraná, FERREIRA et al.15 detected subtype B in 52.6% of cases, followed by subtype C in 29.8%, mosaic BC in 14%, CF in 1.8%, and F in 1.8%. Subtype C was the most prevalent (29%) in a study conducted in 2008 in Porto Alegre, Rio Grande do Sul. Subtype B was detected in 22.6% of the patients, followed by CRF_31BC in 23.4%, subtype F in 10% and other mosaic forms in 5.5%13.
In the present study, the frequency of recombinant forms was 27.1%: B/F (12.5%), B/C (10.4%) and F/C (2.1%), i.e., a relatively high proportion. Similarly high proportions have been documented in studies7 in other areas of Brazil. This probably indicates that different subtypes have co-circulated for some time, especially subtypes B and C in our area, with no segregation of subtype by risk group. This theoretically increases the probability of co-infection and occasional recombination of different subtypes. Our study assessed mostly heterosexual cases, and our sample was limited, so it is not possible to confirm this hypothesis. Larger and appropriately designed studies are necessary to evaluate this issue.
The number of CD4+ T cells showed no relationship with the various subtypes, and the differences observed in the viral load may be due to intra-patient variations in infected individuals. Therefore, the small number of samples in our study does not allow an adequate assessment of this problem. More studies are needed to clarify this important issue. The follow-up of these patients will assess whether the decrease in CD4+ T cells and the need for treatment may help to better understand this issue.
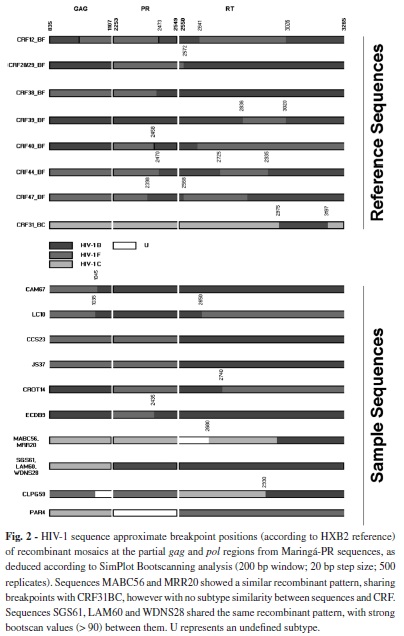
None of the recombinant forms showed a clear phylogenetic relationship or breakpoint structure suggestive of the known CRFs. Three of the recombinants (SGS61, LAM60 and WDNS28) have a similar breakpoint structure, cluster in the phylogenetic tree, and may represent a new CRF. The same was observed with two sequences of a different pattern (MABC56 and MRR20) as shown in Figure 2. To confirm this, epidemiological information, as well as a nearly full-length sequence would be necessary to assign a new CRF to those isolates.
The mutations in the SDRM list were selected as indicators of resistance transmission according to the following criteria: (i) recognized as the cause of or as contributing to resistance, (ii) non-polymorphisms in untreated patients and (iii) applicable to all HIV-1 subtypes17. The prevalence of ARV-transmitted resistance observed in this study (4.2%) was comparable to that reported by SPRINZ et al.34, who investigated 387 treatment-naïve patients from 13 Brazilian cities (Ribeirão Preto, Santo André, Santos, Nova Iguaçu, Rio de Janeiro, Belo Horizonte, Curitiba, Florianópolis, Salvador, Brasília, Campinas, Porto Alegre and São Paulo) and found that 6.7% of the mutations conferred resistance to ARVs. The prevalence of transmitted resistance according to drug class was 1% for PIs, 4.4% to NNRTIs and 1.3% to NRTIs in that study. In another evaluation, FERREIRA et al.15 observed a similar resistance rate of 8.8% (7% to NNRTIs and 1.8% to NRTIs).
Among mutations in the RT gene, the K219N mutation identified in one patient is associated with low-level resistance to the NRTIs zidovudine (AZT) and stavudine (D4T). This mutation was detected in the recombinant isolate Fgag/FPR/BRT. Related mutations were found in treatment-naïve patients from Venezuela, for example, K219R in a recombinant virus Cgag/BRT/CENV 9 and K219Q, a thymidine-analog mutation2.
According to the Stanford algorithm17, mutation V118I, which was found in three patients in this study, causes low-level resistance to lamivudine (3TC) and possibly to other NRTIs in the presence of mutation E44D and/or thymidine-analog mutations21,40. However, according to the Stanford algorithm, this mutation may reduce the activity of 3TC, emtricitabine (FTC), AZT, D4T, didanosine (DDI) and tenofovir (TDF), as observed in approximately 2% of treatment-naïve patients.
Mutations that confer resistance to PIs, V82L and L90M were detected in the same patient. In the study of YAOTSÈ et al.43, the mutation L90M in the protease gene was found to confer resistance to the PIs nelfinavir (NFV), indinavir (IDV) and saquinavir (SQV). This mutation was also observed in the present study. On the other hand, mutation V82L is a rare mutation selected by tipranavir (TPV), which reduces susceptibility to the drug. The effect of this mutation on other PIs has not been characterized1.
The minor mutation L10I/V is associated with resistance to most PIs when other mutations are present. This mutation is detected in 5-10% of untreated patients according to the Stanford database, and reduces the activity of the PIs atazavir/ritonavir (ATV/r), fosamprenavir/r (FPV/r), IDV/r, lopinavir/r (LPV/r), NFV and SQV/r. The L10I/V polymorphism is a mutation that is weakly associated with a reduced response of the virus to some PIs26. The combination of minor mutations M36I and L10I/V has been suggested to result in the subsequent development of mutation L90M, causing virological failure and some level of resistance to all PIs29,36.
Theoretically, patients with a recent infection are more prone to harbor variants of resistant HIV strains than are chronically infected treatment-naïve patients. This can be explained by the fact that wild-type strains become predominant in the absence of selective pressure from a drug, when individuals are infected with both wild-type and mutant viral strains1. However, drug-resistance mutations can be identified in patients with chronic infections, demonstrating the lack of complete reversal in all cases and the persistence of these strains in the plasma of treatment-naïve patients for several years6,18,23,30. In addition, the possibility of transmission of more complex patterns should be taken into account, although they are not detected in treatment-naïve patients because of the reversal of mutations or because of the low sensitivity of the drug-resistance test in detecting variants present in smaller numbers30. In the present study, although no patients with recent infection could be identified, resistance mutations were detected in patients who had been diagnosed more than one year previously.
European protocols recommend resistance testing for all treatment-naïve patients with acute or recent infections, whereas chronically infected patients who require treatment should be tested if the suspicion of resistance is high or if the prevalence of resistance in the population exceeds 10%1,41. In Brazil, resistance testing is recommended only for patients undergoing antiretroviral treatment who present virological failure5. The low prevalence of resistance mutations in our population does not support a recommendation of testing all cases prior to initiating therapy in our area.
In conclusion, HIV-1 subtypes B (39.6%) and C (25%) were the most prevalent in Maringá, with a rather high prevalence of recombinants. If one considers sequences with one or more segments of subtype C, the proportion of B and C is more comparable. Further studies should be conducted in the region to monitor the evolution of the epidemic, to evaluate the trends both in subtype distributions and in the antiretroviral-resistance mutation levels.
Sequence Data: The acession numbers of 142 HIV-1 sequences obtained in this study are as follows: HM752101 - HM752242.
ACKNOWLEDGMENTS
We thank the Serviço de Atendimento Especializado em DST/AIDS of the city of Maringá for permitting this study, the Laboratory of Retroviruses, Instituto Adolfo Lutz, for help with the analyses, and the Laboratory of Retrovirology, Universidade Federal de São Paulo, for help with the interpretation and analysis of the results.
AUTHOR DISCLOSURE
There are no competing financial interests.
Received: 6 September 2011
Accepted: 3 May 2012
- 1. Bakhouch K, Oulad-Lahcen A, Bensghir R, Blaghen M, Elfilali KM, Ezzikouri S, et al. The prevalence of resistance-associated mutations to protease and reverse transcriptase inhibitors in treatment-naïve (HIV-1)-infected individuals in Casablanca, Morocco. J Infect Dev Ctrs. 2009;3:380-91.
- 2. Bouchard M, Masquelier B, Moreno M, Deibis L, Pérez GE, Fleury H, et al. HIV type 1 drug resistance among naïve patients from Venezuela. AIDS Res Hum Retroviruses. 2007;23:482-5.
-
3Brasil. Ministério da Saúde. Boletim epidemiológico AIDS/DST: 27ª a 52ª semanas epidemiológicas - julho a dezembro de 2009 e 01ª a 26ª semanas epidemiológicas - janeiro a junho de 2010, ano VII, no 01. [Cited: 2011 Oct 17]. Available from: www.aids.gov.br
- 4. Brasil. Ministério da Saúde. Departamento de DST, Aids e Hepatites Virais. Monitoraids. Secretaria de Vigilância em Saúde. 2011. [Cited: 2011 Oct 16]. Available from: http://sistemas.aids.gov.br/monitoraids/
-
5Brasil. Ministério da Saúde. Interpretação Brasileira de Genotipagem - Algoritmo Brasileiro (Versão 10: Outubro/2009). Secretaria de Vigilância em Saúde. [Cited: 2010 Jan 22]. Available from: http://www.aids.gov.br/data/Pages/LUMIS1F16A8CFPTBRIE.htm
- 6. Brenner BG, Routy JP, Petrella M, Moisi D, Oliveira M, Detorio M, et al. Persistence and fitness of multidrug-resistant human immunodeficiency virus type 1 acquired in primary infection. J Virol. 2002;76:1753-61.
- 7. Brígido LF, Nunes CC, Oliveira CM, Knoll RK, Ferreira JL, Freitas CA, et al. HIV type 1 subtype C and CB Pol recombinants prevail at the cities with the highest AIDS prevalence rate in Brazil. AIDS Res Hum Retroviruses. 2007;23:1579-86.
- 8. Brindeiro RM, Diaz RS, Sabino EC, Morgado MG, Pires IL, Brígido LV, et al. Brazilian Network for HIV Drug Resistance Surveillance (HIV-BResNET): a survey of chronically infected individuals. AIDS. 2003;17:1063-9.
- 9. Castro E, Moreno M, Deibis L, de Pérez G, Salmen S, Berrueta L. Trends of HIV-1 molecular epidemiology in Venezuela: introduction of subtype C and identification of a novel B/C mosaic genome. J ClinVirol. 2004;32:257-8.
- 10. De Medeiros LB, Lacerda HR, Cavalcanti AM, De Albuquerque M de-F. Primary resistance of human immunodeficiency virus type 1 in a reference center in Recife, Pernambuco, Brazil. Mem Inst Oswaldo Cruz. 2006;101:845-9.
- 11. Dean AG, Sullivan KM, Soe MM. OpenEpi: Open Source Epidemiologic Statistics for Public Health, Version 2.3. [Cited: 2009 March 9]. Available from: www.openepi.com
- 12. Delwart EL, Shpaer EG, Louwagie J, McCutchan FE, Grez M, Rubsamen-Waigmann H, et al. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science.1993,262:1257-61.
- 13. Dias CF, Nunes CC, Freitas IO, Lamego IS, Oliveira IMR, Gilli S, et al. High prevalence and association of HIV-1 non-B subtype with specific sexual transmission risk among antiretroviral naive patients in Porto Alegre, Brazil. Rev Inst Med Trop Sao Paulo. 2009;51:191-6.
- 14. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792-7.
- 15. Ferreira JLP, Thomaz M, Rodrigues R, Harrad D, Oliveira CM, Oliveira CAF, et al. Molecular characterization of newly identified HIV-1 infections in Curitiba, Brazil: preponderance of clade C among males with recent infections. Mem Inst Oswaldo Cruz. 2008;103:800-8.
- 16. Frenkel LM, Wagner LE 2nd, Atwood SM, Cummins TJ, Dewhurst S. Specific, sensitive, and rapid assay for human immunodeficiency virus type 1 pol mutations associated with resistance to zidovudine and didanosine. J Clin Microbiol. 1995;33:342-7.
- 17. Gifford R, Oliveira T, Rambaut A, Myers RE, Gale CV, Dunn D, et al. Assessment of automated genotyping protocols as tools for surveillance of HIV-1 genetic diversity. AIDS. 2006;20:1521-9.
- 18. Gonzalez CR, Alcade R, Nishiya A, Barreto CC, Silva FES, Almeida A, et al. Drug resistance among chronic HIV-1 infected patients naive for use of anti-retroviral therapy in São Paulo city. Virus Res. 2007;129:87-90.
- 19. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95-8.
- 20. Inocencio LA, Pereira AA, Sucupira MC, Fernandez JCC, Jorge CP, Souza DFC, et al. Brazilian Network for HIV Drug Resistance Surveillance: a survey of individuals recently diagnosed with HIV. J Int AIDS Soc. 2009;12(20).
- 21. Johnson VA, Brun-Vezinet F, Clotet B, Conway B, Kuritzkes DR, Pillay D, et al. Update of the drug resistance mutations in HIV-1: Fall 2005. Top HIV Med. 2005;13:125-31.
- 22. Kozal MJ, Shah N, Shen N, Yang R, Fucini R, Merigan TC, et al. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat Med. 1996;2:753-9.
- 23. Little SJ, Dawson K, Hellmann NS, Richmann DD, Frost SDW. Persistence of transmitted drug-resistant virus among subjects with primary HIV infection deferring antiretroviral therapy. Antivir Ther. 2003;8:129.
- 24. Locateli D, Stoco PH, Queiroz ATL, Alcântara LCJ, Ferreira LGE, Zanetti CR, et al. Molecular epidemiology of HIV-1 in Santa Catarina State confirms increases of subtype C in Southern Brazil. J Med Virol. 2007;79:1455-63.
- 25. Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152-60.
- 26. Mintsa-Ndong A, Caron M, Plantier J-C, Makuwa M, Le Hello S, Courgnaud V, et al. High HIV type 1 prevalence and wide genetic diversity with dominance of recombinant strains but low level of antiretroviral drug-resistance mutations in untreated patients in Northeast Gabon, Central Africa. AIDS Res Hum Retroviruses. 2009;25:411-8.
- 27. Myatt M, Bennett DE. A novel sequential sampling technique for the surveillance of transmitted HIV drug resistance by cross-sectional survey for use in low resource settings. Antivir Ther. 2008;13(Suppl 2):37-48.
- 28. Oliveira T, Deforche K, Cassol S, Salminem M, Paraskevis D, Seebregts C, et al. An automated genotyping system for analysis for HIV-1 and other microbial sequences. Bioinformatics. 2005;21:3797-800.
- 29. Perno CF, Cozzi-Lepri A, Forbici F, Bertoli A, Violin M, Stella Mura M, et al. Minor mutations in HIV protease at baseline and appearance of primary mutation 90M in patients for whom their first protease-inhibitor antiretroviral regimens failed. J Infect Dis. 2004;189:1983-7.
- 30. Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31:298-303.
- 31. Rodrigues R, Vazquez CMP, Colares JK, Custodio RM, Bonásser Filho F, Souza LR, et al. Antiretroviral resistance mutations in human immunodeficiency virus type 1 infected patients enrolled in genotype testing at the Central Public Health Laboratory, São Paulo, Brazil: preliminary results. Mem Inst Oswaldo Cruz. 2005;100:97-102.
- 32. Rozanov M, Plikat U, Chappey C, Kochergin A, Tatusova T. A web-based genotyping resource for viral sequences. Nucl Acids Res. 2004;1:32 (Web Server issue):W654-9.
- 33. Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463-7.
- 34. Sprinz E, Netto EM, Patelli M, Lima JS, Furtado JJD, Eira M, et al. Primary antiretroviral drug resistance among HIV type1-infected individuals in Brazil.AIDS Res Hum Retroviruses. 2009;25:861-7.
-
35StatSoft, Inc. STATISTICA (data analysis software system), version 7.1. [cited 2006]. Available from: www.statsoft.com
- 36. Sucupira MCA, Caseiro MM, Alves K, Tescarollo G, Janini LM, Sabino EC, et al. High levels of primary antiretroviral resistance genotypic mutations and B/F recombinants in Santos, Brazil. AIDS Patient Care STDs. 2007;21:116-28.
- 37. Swofford DL. PAUP* Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland: Sinauer Associates; 1998.
- 38. Tamura K, Dudley J, Nei M, Kumar S. MEGA4. Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596-9.
- 39. Togawa RC, Brigido MM. PHPH: Web based tool for simple electropherogram quality analysis. 1st International Conference on Bioinformatics and Computational Biology - IcoBiCoBi. RibeirãoPreto, May 14-16, 2003. Available from: http://asparagin.cenargen.embrapa.br/phph
- 40. Valle-Bahena OM, Ramos-Jiménez J, Ortiz-López R, Revol A, Lugo-Trampe A, Barrera-Saldaña HA, et al. Frequency of protease and reverse transcriptase drug resistance mutations in naive HIV-infected patients. Arch Med Res. 2006;37:1022-7.
- 41. Vandamme AM, Sonnerborg A, Ait-Khaled M, Albert J, Asjo B, Bacheler L, et al. Updated European recommendations for the clinical use of HIV drug resistance testing. Antiv Ther. 2004;9:829-48.
- 42. Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J Hered. 2001;92:371-3.
- 43. Yaotsè DA, Nicole V, Roch NF, Mireille PD, Eric D, Martine P. Genetic characterization of HIV-1 strains in Togo reveals a high genetic complexity and genotypic drug-resistance mutations in ARV naïve patients. Inf Genet Evol. 2009;9:646-52.
- 44. Yearly S, Kaiser L, Race E, Bru JP, Clavel F, Perrin L. Transmission of antiretroviral-drug-resistant HIV-1 variants. Lancet. 1999;354:729-33.
Correspondence to:
Publication Dates
-
Publication in this collection
25 July 2012 -
Date of issue
Aug 2012
History
-
Received
06 Sept 2011 -
Accepted
03 May 2012