Abstract
INTRODUCTION
Salmonella enterica serovar Panama belongs to the D1 serogroup and is frequently associated with nontyphoidal salmonellosis in humans. This study aimed to characterize isolates collected from Northeast Brazil by phenotypic and molecular methods.
METHODS
Forty four S. Panama strains were examined for antimicrobial susceptibility, virulence genes, and pulsed field gel electrophoresis (PFGE) types.
RESULTS
All strains were susceptible to antibiotics (except for streptomycin), presented classical virulence factors, and could be clustered into four groups and 18 pulsotypes.
CONCLUSIONS
This work calls for continuous surveillance for the emergence of antibiotic resistance and new clones in a geographical area.
Keywords:
Salmonella Panama; Antimicrobial susceptibility; Virulence genes; PFGE typing; Northeast Brazil
Nontyphoidal Salmonella serotypes are some of the most common pathogens involved in food-borne self-limiting gastrointestinal illness worldwide that most commonly do not require antimicrobial therapy. However, these pathogens can also lead to life-threatening systemic infections such as sepsis, meningitis, arthritis, and osteomyelitis, which require specific chemotherapy11. Tsai KS, Yang YJ, Wang SM, Chiou CS, Liu CC. Change of serotype pattern of Group D non-typhoidal Salmonella isolated from pediatric patients in southern Taiwan. J Microbiol Immunol Infect. 2007;B40(3):234-9.. Studies have suggested that previous occurrence of gastroenteritis by Salmonella group D in children is a risk factor for bacteremia22. Yang YJ, Huang MC, Wang SM, Wu JJ, Cheng CP, Liu CC. Analysis of risk factors for bacteremia in children with nontyphoidal Salmonella gastroenteritis. Eur J Clin Microbiol Infect Dis. 2002;21(4):290-3.. S. enterica serovar Panama belongs to group D1 and has been isolated from humans, foods, domestic and wild animals, and environmental sources. In humans, S. Panama usually causes gastroenteritis, but tends to cause invasive illnesses in some hosts11. Tsai KS, Yang YJ, Wang SM, Chiou CS, Liu CC. Change of serotype pattern of Group D non-typhoidal Salmonella isolated from pediatric patients in southern Taiwan. J Microbiol Immunol Infect. 2007;B40(3):234-9.,22. Yang YJ, Huang MC, Wang SM, Wu JJ, Cheng CP, Liu CC. Analysis of risk factors for bacteremia in children with nontyphoidal Salmonella gastroenteritis. Eur J Clin Microbiol Infect Dis. 2002;21(4):290-3..
The ability of Salmonella spp. to cause disease can be attributed to an array of virulence genes located in the chromosomes or in large virulence-associated plasmids33. Rowlands REG, Ristori CA, Ikuno AA, Barbosa ML, Jakabi M, Franco BD. Prevalence of drug resistance and virulence features in Salmonella spp. isolated from foods associated or not with salmonellosis in Brazil. Rev Inst Med Trop Sao Paulo. 2014;56(6):461-7.. Several factors related to the virulence of Salmonella spp. have been described, including the presence of fimbriae and flagella, mobility, ability to invade and replicate in epithelial cells, resistance to complement action, and the production of enterotoxin, cytotoxin, and endotoxin. Some of these classic virulence factors are encoded by invA, fimA, agfA, sfbA, phoP/Q, slyA, spvC, and fliC genes. Many of these virulence genes of Salmonella spp. are found on a “Salmonella pathogenicity island” (SPI), which are genetic elements on the chromosomes acquired by horizontal gene transfer. The acquisition of SPIs is considered an important virulence trait for different Salmonella serotypes, allowing colonization at specific regions and increasing the capacity to pass through host barriers44. Kingsley RA, Baumler AJ. Host adaptation and the emergence of infectious diseases: the Salmonella paradigm. Mol Microbiol. 2000;36(5):1006-14..
Few studies have examined the virulence genes in S. Panama55. Soto SM, Guerra B, Del Cerro A, González-Hevia MA, Mendoz MC. Outbreaks and sporadic cases of Salmonella serovar Panama studied by DNA fingerprinting and antimicrobial resistance. Int J Food Microbiol. 2001;71(1):35-43.,66. Del Cerro A, Soto SM, Mendoza MC. Virulence and antimicrobial-resistance gene profiles determined by PCR-based procedures for Salmonella isolated from samples of animal origin. Food Microbiol. 2003;20(4):431-8.. A study with strains isolated from animals in Spain demonstrated that some virulence profiles were serovar-specific and that S. Panama harbored all the virulence genes tested such as invE/A, phoP/Q, stn, iroB, slyA, hin/H2, and agfA66. Del Cerro A, Soto SM, Mendoza MC. Virulence and antimicrobial-resistance gene profiles determined by PCR-based procedures for Salmonella isolated from samples of animal origin. Food Microbiol. 2003;20(4):431-8.. Another study, also carried out in Spain, with isolates from different sources, showed that all the strains of S. Panama examined were negative for spvC, which is a plasmid-located virulence gene, but were positive for invA, phoP, stn, and slyA, which are chromosomally located genes55. Soto SM, Guerra B, Del Cerro A, González-Hevia MA, Mendoz MC. Outbreaks and sporadic cases of Salmonella serovar Panama studied by DNA fingerprinting and antimicrobial resistance. Int J Food Microbiol. 2001;71(1):35-43..
The worldwide emergence of multidrug-resistant Salmonella strains has been described11. Tsai KS, Yang YJ, Wang SM, Chiou CS, Liu CC. Change of serotype pattern of Group D non-typhoidal Salmonella isolated from pediatric patients in southern Taiwan. J Microbiol Immunol Infect. 2007;B40(3):234-9.,77. Hur J, Jawale C, Lee JH. Antimicrobial resistance of Salmonella isolated from food animals: A review. Food Res Int. 2012;45(2):819-30.,88. Matias CAR, Pereira IA, de Araújo MDS, Santos AFM, Lopes RP, Christakis S, et al. Characteristics of Salmonella spp. isolated from wild birds confiscated in illegal trade markets, Rio de Janeiro, Brazil. Biomed Res Int. 2016;2016:1-7.and these strains are usually isolated from patients with bacteremia and other complications11. Tsai KS, Yang YJ, Wang SM, Chiou CS, Liu CC. Change of serotype pattern of Group D non-typhoidal Salmonella isolated from pediatric patients in southern Taiwan. J Microbiol Immunol Infect. 2007;B40(3):234-9.. S. Panama isolates from various origins exhibit antimicrobial resistance, and multidrug-resistant strains are common. In addition, S. Panama infections frequently recur88. Matias CAR, Pereira IA, de Araújo MDS, Santos AFM, Lopes RP, Christakis S, et al. Characteristics of Salmonella spp. isolated from wild birds confiscated in illegal trade markets, Rio de Janeiro, Brazil. Biomed Res Int. 2016;2016:1-7.,99. Ribeiro VB, Andrigheto C, Bersot LS, Barcellos V, Reis ER, Destro MT. Serological and genetic diversity amongst Salmonella strains isolated in a salami processing line. Braz J Microbiol. 2007;38(1):178-82.,1010. Boonkhot P, Tadee P, Patchanee P. Serodiversity and Antimicrobial Resistance Profiles of detected Salmonella on swine production chain in Chiang Mai and Lamphun, Thailand. Acta Sci Vet. 2015;43:1263..
The application of pulsedfield gel electrophoresis (PFGE) for assessing genetic diversity of Salmonella isolates and the clonal transmission of isolates has been reported in both the food and livestock industries. In Brazil, some studies have demonstrated the occurrence of S. Panama mainly in water, poultry and swine meat, in foods such as salami, and in the environment, however, there is little information available on the virulence and genetic diversity of S. Panama strains99. Ribeiro VB, Andrigheto C, Bersot LS, Barcellos V, Reis ER, Destro MT. Serological and genetic diversity amongst Salmonella strains isolated in a salami processing line. Braz J Microbiol. 2007;38(1):178-82.,1111. Moura MS, Oliveira RP, Melo RT, Mendonça EP, Fonseca BB, Rossi DA. Genes de virulência e diversidade genética em Salmonella spp. isoladas de amostras de origem suína. Arq Bras Med Vet Zootec. 2014;66(5):1367-75..
The aim of the present study was to characterize S. Panama isolates, using phenotypic and molecular methods. A total of 44 S. Panama isolates obtained from human, animal, and food sources were used in this study (Table 1). These strains were isolated from the Northeastern region of Brazil between 2001 and 2008 and stored at the Enterobacteria Collection of the National Reference Laboratory of Enteric Diseases (NRLED), Oswaldo Cruz Institute, Rio de Janeiro, Brazil. At the time of this study, all the S. Panama strains present in the collection were analyzed. The phenotypic identification of the isolates was confirmed by standard biochemical laboratory methods. Serological tests were performed by slide agglutination1212. (Secretaria de Vigilância em Saúde). Manual Técnico de Diagnóstico Laboratorial da Salmonella spp. Laboratório de Referência Nacional de Enteroinfecções Bacterianas. Fundação Oswaldo Cruz. Ministério da Saúde, 2011. 60p.. Susceptibility to nalidixic acid, ampicillin, ciprofloxacin, chloramphenicol, spectinomycin, streptomycin, nitrofurantoin, tetracycline, and sulfonamides was determined by the disk-diffusion method1313. Clinical Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing: 25th informational supplement. Wayne, PA: CLSI; CLSI document M100-S25. 2015.. Virulence genes were detected by PCR using the primers listed in Table 2. Assays were performed in a final volume of 25 μl consisting 2.5 μl template Salmonella DNA that was added to 17.5 µl of ultrapure water (MilliQ), 2.5 µl of 10X PCR buffer (Promega Co., Madison, WI, USA), 0.75 µl of MgCl2 (50 mM) [Promega Co., Madison, WI, USA], 0.5 µl of dNTP mix 10 mM (dATP, dGTP, dCTP, and dTTP) [Biotools B&M Labs, S.A.], 0.5µl of each primer (5 pmol/µl), and 0.25 µl of 1U/µl Taq DNA polymerase (Biotools B&M Labs, S.A.). Amplifications were carried out using the following program: a hot start cycle of 94°C for 2min, followed by 30 cycles of 94°C for30 s, 56°C for 30 s, and a final extension step of 72°C for 2min. Aliquots of 10 µl of the amplification products were analyzed by electrophoresis on 2% agarose (Invitrogen) gels. Gels were stained with ethidium bromide (0.5 µg/ml) and visualized with UV light using a DNR Bio-Imaging Systems (BioAmerica, Miami, Florida, USA) with a Gel Capture software version 5.0. PFGE was performed according to the standardized Salmgene and PulseNet protocol1414. Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, et al. Standardization of Pulsed-Field Gel Electrophoresis Protocols for the Subtyping of Escherichia coli 0157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3(1):59-67.. S. enterica serotype Braenderup H9812 was used as the molecular size marker in the PFGE experiment. DNA patterns were analyzed using BioNumerics software (V 4.1, Applied Maths, Kortrijk, Belgium). Algorithms available within the program were used to compare patterns. Dendrograms were produced using the Dice coefficient and the unweighted pair group method with arithmetic averages (UPGMA), with a 1% tolerance limit and 1% optimization. Isolates with more than 80% similarity were clustered as highly genetically related and were placed into dendrogram branches numbered 1 to 14. Genetically indistinguishable isolates (similarities less than 80%) were assigned a capital letter: A, B, C, or D.
The characteristics of the 44 S. Panama strains included in the study are presented in Table 1. Among the 14 human strains, 12 were obtained from the feces of patients with gastroenteritis and two strains were isolated from patients with an invasive infection, both living in Ceará state; strain 1901was isolated from the blood of an adult woman in 2007 and strain 2625 was isolated from the cerebrospinal fluid (CSF) of a 4-month-old child in 2008. The other 30 strains were isolated from food (n=24), animals (n=4), and water (n=2).
All strains were found to be susceptible to ampicillin, ciprofloxacin, chloramphenicol, nalidixic acid, sulfonamides, tetracycline, and spectinomycin, but only 6.8% of the strains were susceptible to streptomycin. Similar resistance rates to streptomycin have been identified in some other studies1515. Huang SC, Chiu CH, Chiou CS, Yang YJ. Multidrug-resistant Salmonella enterica serovar Panama carrying class 1 integrons is invasive in Taiwanese children. J Formos Med Assoc. 2013;112(5):269-75.,1616. Lee HY, Yang YJ, Su LH, Hsu CH, Fu YM, Chiu CH. Genotyping and antimicrobial susceptibility of Salmonella enterica serotype Panama isolated in Taiwan. J Microbiol Immunol Infect . 2008;41(6):507-12.. In developed countries, antimicrobial drug resistance in nontyphoidal salmonella is commonly related to the use of antimicrobial drugs in animals used as sources of food, but in contrast, in developing countries, the occurrence of resistance in both nontyphoidal and typhoidal Salmonella spp. has been associated with the use of antimicrobials in human medicine. In addition, the occurrence of resistant Salmonella spp. isolates harboring resistance genes in plasmids and integrons can lead to the dissemination of these genes at the inter- or intra-species level. This possibility justifies the need for regular epidemiological surveillance to detect the emergence of antimicrobial-resistant Salmonella strains, which are of great concern worldwide.
The detection of virulence-related genes showed that all 44 isolates were positive for invA (invasion), fimA and agfA (adherence), and sfbA (component of the iron transport system) genes and negative for spvC (motility) and fliC (adherence and invasion) genes. Only one isolate (231/2001) was negative for phoP/Q (intra-macrophage survival),while slyA (a virulence-associated transcriptional regulator) was detected in 12 isolates obtained from food (7), estuarine water (2), and rectal swab from a lizard (3). Salmonella infections show multifactorial pathogenesis and that multiple virulence genes may be involved. Almost all the isolates analyzed in this study showed the presence of invA, fimA, agfA, sfbA, and phoP/Q genes, but not the spvC and fliC genes. These results are similar to those reported in other studies66. Del Cerro A, Soto SM, Mendoza MC. Virulence and antimicrobial-resistance gene profiles determined by PCR-based procedures for Salmonella isolated from samples of animal origin. Food Microbiol. 2003;20(4):431-8.. Only 40% of our isolates contained the slyA gene; this result differs from results of other studies that have detected this gene in the majority of S. Panama isolates. The presence of these virulence genes could play an important role and contribute to the capacity of this serotype to cause disease, including invasive illnesses such as sepsis and meningitis. Experiments carried out using a murine infection model showed that some serotypes of Salmonella could have invasive characteristics; however, S. Panama did not show this ability1717. Helmuth R, Stephan R, Bunge C, Hoog B, Steinbeck A, Bulling E. Epidemiology of virulence-associated plasmids and outer membrane protein patterns within seven common Salmonella serotypes. Infect Immun. 1985;48(1):175-82.. However, another study suggested that S. Panama is more invasive than S. Typhimurium when tested in HEp-2 cells2. Indeed, more studies are needed to elucidate the role of the virulence-related genes in the adherence, colonization, and invasiveness of the S. Panama serotype.
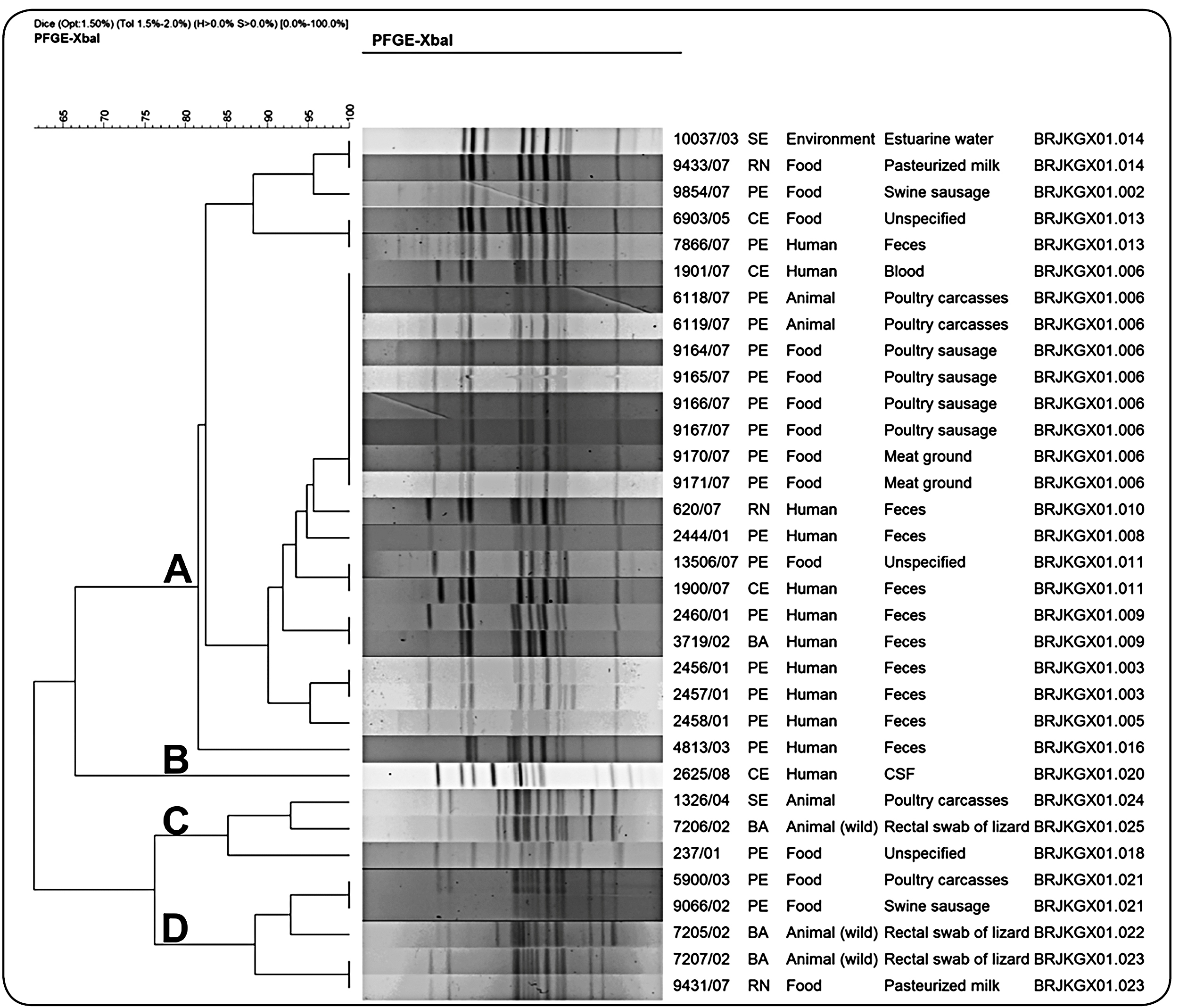
In the present study, PFGE, which is considered the gold standard typing method for Salmonella, was used to assess genetic relatedness (Figure 1). The application of the Dice coefficient and an 80% similarity threshold allowed the identification of 18 pulsotypes that were grouped in four clusters: A (24 isolates, 11 pulsotypes), B (1 isolate, 1 pulsotype), C (3 isolates, three pulsotypes), and D (5 isolates, three pulsotypes). Degradation of DNA was an inhibiting factor when typing the isolates by PFGE, and even with the use of high concentrations of thiourea, only 33 XbaI-digested samples could be typed. Cluster A, which consisted of S. Panama samples collected from humans, food, the environment, and animal origin (during a period of five years from five different states), showed the dispersal ability and persistence of S. Panama. Human isolates (91.0%) were included in this cluster, and the majority showed a similarity of 90%. A more invasive clone was probably the origin of these isolates. In addition, BRJKGX01.011 and BRJKGX01.013 pulsotypes consisted of seemingly unrelated and epidemiologically indistinguishable samples. Cluster B, represented by one human isolate, was collected from cerebrospinal fluid (CSF) in 2008. Three isolates belonging to cluster C isolated in Bahia, Pernambuco, and Sergipe, from 2001 to 2004, showed ≥ 85% similarity. The isolates from Bahia (2002) and Sergipe (2004) were from a poultry carcass and a wild animal (lizard), respectively, and showed 92.5% similarity. Two isolates from Bahia, two from Pernambuco, and one from Rio Grande do Norte belonged to cluster D. Two epidemiologically unrelated isolates (one isolated from a wild animal in Bahia in 2002 and the other from pasteurized milk in Rio Grande do Norte in 2007) were placed together and one indistinguishable PFGE pulsotype was observed.
profiles of 33 XbaI-digested Salmonella Panama isolates showing state of origin, source of the isolate, and pulsotypes.
Very little information has been published on the molecular epidemiology of S. Panama from Brazil99. Ribeiro VB, Andrigheto C, Bersot LS, Barcellos V, Reis ER, Destro MT. Serological and genetic diversity amongst Salmonella strains isolated in a salami processing line. Braz J Microbiol. 2007;38(1):178-82.. A study with 110 isolates of S. Panama, including isolates from the environment (n=84), humans (n=16), food (n=5), and wild animals (n=5) obtained from different cities in the state of Pará, Brazil reported that 81% of the isolates were typeable by PFGE and a high genetic diversity with 54 distinct profiles and 16 clones was detected. The presence of multiple PFGE profiles was also detected among S. Panama isolated from salami99. Ribeiro VB, Andrigheto C, Bersot LS, Barcellos V, Reis ER, Destro MT. Serological and genetic diversity amongst Salmonella strains isolated in a salami processing line. Braz J Microbiol. 2007;38(1):178-82.in the state of Parana, Brazil. In a study conducted in Taiwan, to investigate the changing serotypes, genotypes and antibiotic resistance of 81 isolates from group D Salmonella infection, S. enterica serotype Enteritidis was the most common serotype (80%), followed by S. Panama (7%); but S. Panama was associated with a higher rate of bacteremia and antimicrobial resistance than S. Enteritidis. In addition, a PFGE study showed a single genotype of S. Enteritidis and diverse genotypes of S. Panama11. Tsai KS, Yang YJ, Wang SM, Chiou CS, Liu CC. Change of serotype pattern of Group D non-typhoidal Salmonella isolated from pediatric patients in southern Taiwan. J Microbiol Immunol Infect. 2007;B40(3):234-9.. In another study, also conducted in Taiwan, antimicrobial susceptibility testing and molecular typing were performed on nine clinical S. Panama isolates using PFGE. Three predominant PFGE types with six subtypes were found among these isolates1616. Lee HY, Yang YJ, Su LH, Hsu CH, Fu YM, Chiu CH. Genotyping and antimicrobial susceptibility of Salmonella enterica serotype Panama isolated in Taiwan. J Microbiol Immunol Infect . 2008;41(6):507-12..
This study highlighted the importance of continuous surveillance for detecting the antibiotic resistance profiles and the introduction of new clones into a geographical area, information that can be useful for clinicians in choosing appropriate antibiotics for treating severe group D Salmonella infections and should be considered when control programs are planned.
To conclude, our results showed that at the time of this study, the strains of S. Panama circulating in the northeast region of Brazil were consistently collected from both humans and non-human sources and shared similar genotypes within each cluster.
ACKNOWLEDGEMENTS
The authors thank LACEN (Central Laboratory of Public Health) of the Ceará, Pernambuco, Bahia, Sergipe, and Rio Grande do Norte states for providing Salmonella strains.
REFERENCES
-
1Tsai KS, Yang YJ, Wang SM, Chiou CS, Liu CC. Change of serotype pattern of Group D non-typhoidal Salmonella isolated from pediatric patients in southern Taiwan. J Microbiol Immunol Infect. 2007;B40(3):234-9.
-
2Yang YJ, Huang MC, Wang SM, Wu JJ, Cheng CP, Liu CC. Analysis of risk factors for bacteremia in children with nontyphoidal Salmonella gastroenteritis. Eur J Clin Microbiol Infect Dis. 2002;21(4):290-3.
-
3Rowlands REG, Ristori CA, Ikuno AA, Barbosa ML, Jakabi M, Franco BD. Prevalence of drug resistance and virulence features in Salmonella spp. isolated from foods associated or not with salmonellosis in Brazil. Rev Inst Med Trop Sao Paulo. 2014;56(6):461-7.
-
4Kingsley RA, Baumler AJ. Host adaptation and the emergence of infectious diseases: the Salmonella paradigm. Mol Microbiol. 2000;36(5):1006-14.
-
5Soto SM, Guerra B, Del Cerro A, González-Hevia MA, Mendoz MC. Outbreaks and sporadic cases of Salmonella serovar Panama studied by DNA fingerprinting and antimicrobial resistance. Int J Food Microbiol. 2001;71(1):35-43.
-
6Del Cerro A, Soto SM, Mendoza MC. Virulence and antimicrobial-resistance gene profiles determined by PCR-based procedures for Salmonella isolated from samples of animal origin. Food Microbiol. 2003;20(4):431-8.
-
7Hur J, Jawale C, Lee JH. Antimicrobial resistance of Salmonella isolated from food animals: A review. Food Res Int. 2012;45(2):819-30.
-
8Matias CAR, Pereira IA, de Araújo MDS, Santos AFM, Lopes RP, Christakis S, et al. Characteristics of Salmonella spp. isolated from wild birds confiscated in illegal trade markets, Rio de Janeiro, Brazil. Biomed Res Int. 2016;2016:1-7.
-
9Ribeiro VB, Andrigheto C, Bersot LS, Barcellos V, Reis ER, Destro MT. Serological and genetic diversity amongst Salmonella strains isolated in a salami processing line. Braz J Microbiol. 2007;38(1):178-82.
-
10Boonkhot P, Tadee P, Patchanee P. Serodiversity and Antimicrobial Resistance Profiles of detected Salmonella on swine production chain in Chiang Mai and Lamphun, Thailand. Acta Sci Vet. 2015;43:1263.
-
11Moura MS, Oliveira RP, Melo RT, Mendonça EP, Fonseca BB, Rossi DA. Genes de virulência e diversidade genética em Salmonella spp. isoladas de amostras de origem suína. Arq Bras Med Vet Zootec. 2014;66(5):1367-75.
-
12(Secretaria de Vigilância em Saúde). Manual Técnico de Diagnóstico Laboratorial da Salmonella spp. Laboratório de Referência Nacional de Enteroinfecções Bacterianas. Fundação Oswaldo Cruz. Ministério da Saúde, 2011. 60p.
-
13Clinical Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing: 25th informational supplement. Wayne, PA: CLSI; CLSI document M100-S25. 2015.
-
14Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, et al. Standardization of Pulsed-Field Gel Electrophoresis Protocols for the Subtyping of Escherichia coli 0157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3(1):59-67.
-
15Huang SC, Chiu CH, Chiou CS, Yang YJ. Multidrug-resistant Salmonella enterica serovar Panama carrying class 1 integrons is invasive in Taiwanese children. J Formos Med Assoc. 2013;112(5):269-75.
-
16Lee HY, Yang YJ, Su LH, Hsu CH, Fu YM, Chiu CH. Genotyping and antimicrobial susceptibility of Salmonella enterica serotype Panama isolated in Taiwan. J Microbiol Immunol Infect . 2008;41(6):507-12.
-
17Helmuth R, Stephan R, Bunge C, Hoog B, Steinbeck A, Bulling E. Epidemiology of virulence-associated plasmids and outer membrane protein patterns within seven common Salmonella serotypes. Infect Immun. 1985;48(1):175-82.
Publication Dates
-
Publication in this collection
18 July 2019 -
Date of issue
2019
History
-
Received
05 July 2018 -
Accepted
01 Mar 2019