Abstract
INTRODUCTION:
In this study, we report a clonal dissemination of carbapenem resistant Acinetobacter baumannii isolates due to the acquisition of blaOXA-23 in a regional hospital located in Brazilian Amazon Region.
METHODS:
The isolates were identified by MALDI-TOF and the carbapenemase-encoding genes were detected by multiplex-PCR. The genetic similarity was investigated by pulsed-field gel electrophoresis (PFGE).
RESULTS:
Only 10 (55.6%) isolates harbored the gene bla OXA-23. PFGE analysis revealed that these isolates belong to a single clone.
CONCLUSIONS:
This dissemination strategy indicates the need for surveillance, adoption of control procedures defined in guidelines, and the careful administration of antimicrobials should be reinforced.
Keywords:
Acinetobacter baumannii; Carbapenem resistance; Carbapenemase;
bla
OXA-23
; Gram-negative bacilli
Acinetobacter baumannii is an opportunistic pathogen with several virulence factors associated with several outbreaks worldwide, especially among intensive care unit (ICU) patients with severe underlying diseases11. Founou RC, Founou, LL, Essack SY. Clinical and economic impact of antibiotic resistance in developing countries: A systematic review and meta-analysis. PLoS One. 2017;12(12):1-18.,22. Chhatwal KWP, Vonbe RP. Nosocomial outbreaks caused by Acinetobacter baumannii and Pseudomonas aeruginosa: Results of a systematic review. Am J Infect Control. 2018;46(6):643-8.. These infections result in high mortality rates and increased treatment costs11. Founou RC, Founou, LL, Essack SY. Clinical and economic impact of antibiotic resistance in developing countries: A systematic review and meta-analysis. PLoS One. 2017;12(12):1-18.,22. Chhatwal KWP, Vonbe RP. Nosocomial outbreaks caused by Acinetobacter baumannii and Pseudomonas aeruginosa: Results of a systematic review. Am J Infect Control. 2018;46(6):643-8.. Approximately 700,000 deaths worldwide are associated with multidrug-resistant (MDR) microorganisms. By 2050, the estimated global economic losses could reach 60 to 100 trillion dollars if multidrug resistance is not controlled33. Sutherland N, Barber S. O'Neill. Review into Antibiotic Resistance. House of Commons Library, F1, Issue 1. 2017. Available from: https://research briefings .parliament.uk/ Research-Briefing/Summary/CDP-2017-0074.
https://research briefings .parliament.u...
.
The worldwide emergence and dissemination of MDR bacteria, such as A. baumannii, led the World Health Organization (WHO) to gather global leaders at the United Nations (UN) General Assembly meeting in 2016 to commit them to fight against antimicrobial resistance44. World Health Organization (WHO). Chemotherapy of leprosy for control programmes. Technical Report Series 675. Geneva: WHO; 1982. 36 p..
In infections caused by A. baumannii, genes encoding resistance to multiple broad-spectrum antimicrobials are commonly detected, including carbapenem resistance genes55. Castilho SRA, Godoy CSM, Guilarde AO, Cardoso JL, André MCP, Junqueira KAP, et al. Acinetobacter baumannii strains isolated from patients in intensive care units in Goiânia, Brazil: Molecular and drug susceptibility profiles. PLoS One . 2017;12(5):1-13.. A study conducted by the SENTRY Antimicrobial Surveillance Program demonstrated that carbapenem resistance in Brazilian A. baumannii isolates increased by approximately 60% compared to resistance between the periods of 1977-1999 (12.6%) and 2008-2010 (71.4%)66. Gales AC, Castanheira M, Jones RN, Sader HS. Antimicrobial resistance among Gram-negative bacilli isolated from Latin America: results from SENTRY Antimicrobial Surveillance Program (Latin America, 2008-2010). Diagn Microbiol Infect Dis. 2012;73(4):354-60..
In addition, it should be noted that environmental and patient colonization by MDR A. baumannii (MDR-AB) is a risk factor for the dissemination of this pathogen among patients and for the subsequent development of infections77. Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol. 2018;16(2):91-102.. This bacterium remains viable for long periods in the environment, tolerates desiccation and is able to survive on inanimate dry surfaces for several months. All these conditions favor its rapid dissemination by cross-contamination in hospital environments77. Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol. 2018;16(2):91-102..
Hence, MDR-AB isolates are associated with high mortality rates, costs, and dissemination among hospitalized patients. Studying MDR-AB dissemination is important for implementing effective infection control/colonization measures, breaking the epidemiological chain of transmission of this microorganism, mitigating rates of bacterial resistance, reducing morbidity and mortality, and improving the quality of healthcare.
Considering these factors and the lack of research on this topic in the Brazilian Amazon region, the current study aimed to determine the phenotypic and genotypic characteristics of MDR-AB isolates, to detect the presence of carbapenemases, and to demonstrate the genetic similarity among carbapenemase-producing isolates at a tertiary referral hospital in the Amazon.
A cross-sectional descriptive study was performed between September 2017 and February 2018 at a regional public hospital in the southeast region of Pará state (Brazil). This is a tertiary referral hospital with different medical specialties and with pediatric and adult intensive care units, particularly for nephrology, with kidney transplantation and renal replacement therapy services. The hospital treats an estimated population of 541,000 inhabitants and is located in the Amazon biome.
A total of 18 bacterial isolates were recovered from patients undergoing treatment at the regional hospital and diagnosed with a hospital-acquired infection (HAI) or with colonization caused by A. baumannii resistant to imipenem and meropenem were included in the study. Only one isolate per patient was analyzed. The isolates were obtained from clinical samples including tracheal secretions, blood cultures, postoperative wound swabs, catheter tips, and inguinal swabs.
Antimicrobial susceptibility was performed using the disk diffusion test and the results were interpreted according to the Brazilian Committee on Antimicrobial Susceptibility Testing (BrCAST) guidelines88. Brazilian Committee on Antimicrobial Susceptibility Testing (BrCAST). Tabelas de pontos de corte para interpretação de CIMs e diâmetros de halos. Versão. 2020;8:2. for the following antimicrobials: piperacillin-tazobactam, ceftazidime, cefotaxime, cefepime, gentamicin, amikacin, tetracycline, ciprofloxacin, levofloxacin, trimethoprim sulfamethoxazole, imipenem, meropenem, and aztreonam.
The minimum inhibitory concentration (MIC) of carbapenems was defined using the ETEST® strip (bioMérieux) for isolates resistant to imipenem and also using the disk diffusion test for meropenem. MIC of polymyxin B (Sigma-Aldrich, St. Louis, MO, USA) was determined using the broth microdilution susceptibility test (Basingstoke, UK). The concentrations tested ranged from 0.125 to 64 µg/mL and the bacteria were considered resistant when the MIC values were ≥ 4 µg/mL88. Brazilian Committee on Antimicrobial Susceptibility Testing (BrCAST). Tabelas de pontos de corte para interpretação de CIMs e diâmetros de halos. Versão. 2020;8:2..
MIC for tigecyclines was determined using the MicroScan AlkAway® 96 plus (MIC panel type 40, Beckman Coulter, West Sacramento, USA) and interpreted according to the thresholds set by the Food and Drug Administration (FDA) (sensitive ≤2 µg/mL and resistant ≥8 µg/mL; https://www.accessdata.fda.gov/drugsatfda_docs). The standard strain E. coli ATCC 35218 was used as a control. The kits Carbapenembac™ and Carbapenembac-Metalo™ (PROBAC®) were used for phenotypic detection of carbapenemases and confirmation of positive isolates, respectively.
The identification of all positive isolates in the phenotypic test for carbapenemases (n = 10) was confirmed by MALDI-TOF. For genotyping the isolates, chromosomal DNA was extracted from 3-5 colonies after boiling for 15 min and centrifuged at 12,000 rpm at room temperature. PCR was performed in a final volume of 25 µL of master mix (TopTaq®- QIAGEN), containing 25 ng of DNA and 0.5 µL of each primer99. Brasiliense Danielle, Cayô Rodrigo, Streling Ana Paula, Nodari Carolina S, Barata Rafael R, Lemos Poliana S, et al. Diversity of metallo-β-lactamase-encoding genes found in distinct species of Acinetobacter isolated from the Brazilian Amazon Region. Mem Inst Oswaldo Cruz [Internet]. 2019;114(1):1-6.. Primers for the following genes were used: bla VIM, bla SPM, bla GIM, bla IMP, bla SIM, bla NDM, bla KPC, bla OXA-23, bla OXA-24, bla OXA-51, bla OXA-58, and bla OXA-14399. Brasiliense Danielle, Cayô Rodrigo, Streling Ana Paula, Nodari Carolina S, Barata Rafael R, Lemos Poliana S, et al. Diversity of metallo-β-lactamase-encoding genes found in distinct species of Acinetobacter isolated from the Brazilian Amazon Region. Mem Inst Oswaldo Cruz [Internet]. 2019;114(1):1-6..
Genetic similarity between A. baumannii isolates was determined by pulsed-field gel electrophoresis (PFGE), using the molecular weight marker Lambda PFGE Ladder® (GelSyringe ™). Isolates were sent to laboratory of Alerta at the Federal University of São Paulo (UFSP) for molecular typing. For this purpose, bacterial suspensions were digested with the restriction enzyme ApaI (Uniscience, Miami, USA) and the DNA fragments were separated by 1% agarose gel electrophoresis (Invitrogen, Eragny, France) in 0.5X TBE buffer (Tris base, boric acid and EDTA in distilled water). Electrophoresis was performed using the CHEF-DR II system (Bio-Rad Laboratories, California, USA) at 14 ºC, using 200 volts (6 V/cm) electric current with an initial switch time of 5 s and a final switch time of 35 s for 19 hrs. The gel was stained with UniSafe Dye® (Uniscience, Miami, USA) and photographed under ultraviolet light. PFGE-stained photos and DNA fragments were examined using BioNumerics software version 5.0 (Applied Maths, Kortrijk, BE)99. Brasiliense Danielle, Cayô Rodrigo, Streling Ana Paula, Nodari Carolina S, Barata Rafael R, Lemos Poliana S, et al. Diversity of metallo-β-lactamase-encoding genes found in distinct species of Acinetobacter isolated from the Brazilian Amazon Region. Mem Inst Oswaldo Cruz [Internet]. 2019;114(1):1-6.. The bands were automatically defined by the software and then individually checked by visual comparison. Data were interpreted according to the Sørensen-Dice coefficient1010. Dice LR. Measures of the Amount of Ecologic Association Between Species. Ecology. 1945;26(3):297-302., and the dendrogram was constructed using the unweighted pair group method using arithmetic averages (UPGMA). Tolerance was set at 0.8%, and a similarity threshold of 80% was used to separate isolates into clonal clusters.
Samples were obtained from ICU patients (50%) and ward patients (50%). The isolates were collected from tracheal secretions (50%; n = 9), blood cultures (16.7%; n = 3), postoperative wound swabs (16.7%; n = 3), catheter tips (11.1%; n = 2), and inguinal swabs (5.6%; n = 1). Among the 18 isolates included in the study, the majority of patients were male (61.1%; n = 11), whose ages ranged from 18 to 86 years. The isolates were resistant to almost all antimicrobials tested, and remained susceptible to tigecycline and polymyxin B. As shown in Table1, one isolate was not tested for polymyxin B and tigecycline due to loss of cell viability during the susceptibility test (Table 1).
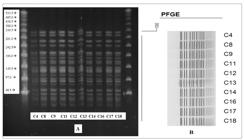
The phenotypic assay showed that all isolates were MDR bacteria, including 10 (55.6%) which were carbapenemase-producing. Three of them (16.7%) were positive for metalo-β-lactamases and seven (83.3%) for serine carbapenemases. However, genotypic testing did not confirm these findings (Table 1). In all 10 isolates characterized as carbapenemase-producing, we found the presence of bla OXA-23 and bla OXA-51 gene. PFGE analysis showed that all isolates which tested positive for carbapenemase belonged to a single clone (Figure 1).
A) PFGE demonstrating similarity among carbapenemase-producing isolates positive for bla-OXA-23. B) A dendrogram representing PFGE profiles of carbapenemase-producing A. baumannii isolates from 10 patients undergoing treatment at a Brazilian Amazon hospital. The identification number of the isolates is found to the right of the profiles.
MDR-AB is more frequently isolated from colonized and/or HAI patients, especially in hospital environments22. Chhatwal KWP, Vonbe RP. Nosocomial outbreaks caused by Acinetobacter baumannii and Pseudomonas aeruginosa: Results of a systematic review. Am J Infect Control. 2018;46(6):643-8.
3. Sutherland N, Barber S. O'Neill. Review into Antibiotic Resistance. House of Commons Library, F1, Issue 1. 2017. Available from: https://research briefings .parliament.uk/ Research-Briefing/Summary/CDP-2017-0074.
https://research briefings .parliament.u...
-44. World Health Organization (WHO). Chemotherapy of leprosy for control programmes. Technical Report Series 675. Geneva: WHO; 1982. 36 p.. These infections occur mainly in patients with severe underlying disease and poor prognosis who are treated with invasive procedures, using broad-spectrum antibiotics, and are admitted to the ICU22. Chhatwal KWP, Vonbe RP. Nosocomial outbreaks caused by Acinetobacter baumannii and Pseudomonas aeruginosa: Results of a systematic review. Am J Infect Control. 2018;46(6):643-8.
3. Sutherland N, Barber S. O'Neill. Review into Antibiotic Resistance. House of Commons Library, F1, Issue 1. 2017. Available from: https://research briefings .parliament.uk/ Research-Briefing/Summary/CDP-2017-0074.
https://research briefings .parliament.u...
-44. World Health Organization (WHO). Chemotherapy of leprosy for control programmes. Technical Report Series 675. Geneva: WHO; 1982. 36 p.. Treating infections caused by MDR-AB is complicated, especially when these bacteria are resistant to all antimicrobials commonly used in clinical practice1111. Genteluci GL, Gomes DBC, Souza MJ, Carvalho KR, Villas BMHS. Emergence of polymyxin B-resistant Acinetobacter baumannii in hospitals in Rio de Janeiro. J Bras Patol Med Lab. 2016;52(2):91-5.. In the present study, all isolates showed a multi-resistance profile, and some showed similar polymyxin B sensitivity rates. This is likely due to extensive and/or inadequate use of these antimicrobials in the treatment of infections caused by gram-negative MDR bacteria1111. Genteluci GL, Gomes DBC, Souza MJ, Carvalho KR, Villas BMHS. Emergence of polymyxin B-resistant Acinetobacter baumannii in hospitals in Rio de Janeiro. J Bras Patol Med Lab. 2016;52(2):91-5.. A study conducted in Rio de Janeiro, Brazil, demonstrated that most isolates (81.5%) were resistant to polymyxins, highlighting the importance of using antimicrobials adequately1111. Genteluci GL, Gomes DBC, Souza MJ, Carvalho KR, Villas BMHS. Emergence of polymyxin B-resistant Acinetobacter baumannii in hospitals in Rio de Janeiro. J Bras Patol Med Lab. 2016;52(2):91-5..
In this study, most of the isolates were susceptible to tigecycline. Although this antimicrobial has been licensed for treating complicated intra-abdominal and skin infections and community-based bacterial pneumonia, it has been widely used off-label to treat many other infections, including those caused by MDR-AB1212. Shin JA, Chang YS, Kim HJ, Kim SK, Chang J, Ahn CM, Byun MK. Clinical Outcomes of Tigecycline in the Treatment of Multidrug-Resistant Acinetobacter baumannii Infection. Yonsei Med J. 2012;53(5):974-84.. However, treatment should be individualized and defined based on the best evidence obtained for combinatorial approaches1212. Shin JA, Chang YS, Kim HJ, Kim SK, Chang J, Ahn CM, Byun MK. Clinical Outcomes of Tigecycline in the Treatment of Multidrug-Resistant Acinetobacter baumannii Infection. Yonsei Med J. 2012;53(5):974-84..
Most A. baumannii isolates in this study were collected from colonized ICU patients. It should be noted that either patient colonization or infection with MDR-AB are important sources of dissemination of resistant strains between hospitals1212. Shin JA, Chang YS, Kim HJ, Kim SK, Chang J, Ahn CM, Byun MK. Clinical Outcomes of Tigecycline in the Treatment of Multidrug-Resistant Acinetobacter baumannii Infection. Yonsei Med J. 2012;53(5):974-84.. One such example was the intercontinental transfer of patients colonized by MDR-AB after repatriation from Tahiti, resulting in a prolonged outbreak77. Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol. 2018;16(2):91-102..
The genes bla OXA-23 and bla OXA-51 were detected in all carbapenemase-producing isolates, similar to what has been described in a previous study, which reported a carbapenem resistance rate of 90%1313. Schuertz KF, Tuon FF, Palmeiro JK, Conte D, Telles JPM, Trevisoli LE, et al. Bacteremia and meningitis caused by OXA-23-producing Acinetobacter baumannii - molecular characterization and susceptibility testing for alternative antibiotics. Braz J Microbiol. 2018;49(s1):199-204.. These genes encode the most common OXA-type carbapenemases that contribute to resistance to imipenem and meropenem in A. baumannii isolates that are endemic in several Brazilian states1313. Schuertz KF, Tuon FF, Palmeiro JK, Conte D, Telles JPM, Trevisoli LE, et al. Bacteremia and meningitis caused by OXA-23-producing Acinetobacter baumannii - molecular characterization and susceptibility testing for alternative antibiotics. Braz J Microbiol. 2018;49(s1):199-204..
The oxacillinase (OXA)-type carbapenemase expression is the most common mechanism of resistance in A. baumannii, but less effective than other enzymatic mechanisms1313. Schuertz KF, Tuon FF, Palmeiro JK, Conte D, Telles JPM, Trevisoli LE, et al. Bacteremia and meningitis caused by OXA-23-producing Acinetobacter baumannii - molecular characterization and susceptibility testing for alternative antibiotics. Braz J Microbiol. 2018;49(s1):199-204.. Nevertheless, oxacillinase-producing isolates are known to be MDR bacteria. Microorganisms expressing blaOXA-23 have high MICs for imipenem and meropenem, but those expressing only bla OXA-51 have a lower MIC due to the reduced hydrolytic activity of OXA-51 for carbapenens55. Castilho SRA, Godoy CSM, Guilarde AO, Cardoso JL, André MCP, Junqueira KAP, et al. Acinetobacter baumannii strains isolated from patients in intensive care units in Goiânia, Brazil: Molecular and drug susceptibility profiles. PLoS One . 2017;12(5):1-13.,1313. Schuertz KF, Tuon FF, Palmeiro JK, Conte D, Telles JPM, Trevisoli LE, et al. Bacteremia and meningitis caused by OXA-23-producing Acinetobacter baumannii - molecular characterization and susceptibility testing for alternative antibiotics. Braz J Microbiol. 2018;49(s1):199-204..
The PFGE pattern demonstrated a high genetic similarity and dissemination of bla OXA-23-encoding A. baumannii strains, matching previous studies on ICU isolates which reported 91.8% and 100% of genetic identity1414. Dettori M, Piana A, Deriu MG, Lo Curto P, Cossu A, Musumeci R, et al. Outbreak of multidrug-resistant Acinetobacter baumannii in an intensive care unit. New Microbiol. 2014:37(2):185-91.,1515. Gokmen T, Akcimen B, Kayar B, Marzi M, Koksal F. The outbreak of Acinetobacter baumannii producing OXA-23 and OXA-51 type carbapenemases in a state hospital. J Exp Clin Med. 2016;33(3):157-61.. This PFGE pattern suggests cross-contamination of A. baumannii isolates related to patient infection or colonization, whose source of dissemination could have been health staff, equipment, or contaminated fomites1414. Dettori M, Piana A, Deriu MG, Lo Curto P, Cossu A, Musumeci R, et al. Outbreak of multidrug-resistant Acinetobacter baumannii in an intensive care unit. New Microbiol. 2014:37(2):185-91.. Ward patients included in the study had been previously admitted to the ICU, which suggests that the ICU was the primary source of cross-contamination of MDR-AB1414. Dettori M, Piana A, Deriu MG, Lo Curto P, Cossu A, Musumeci R, et al. Outbreak of multidrug-resistant Acinetobacter baumannii in an intensive care unit. New Microbiol. 2014:37(2):185-91.,1515. Gokmen T, Akcimen B, Kayar B, Marzi M, Koksal F. The outbreak of Acinetobacter baumannii producing OXA-23 and OXA-51 type carbapenemases in a state hospital. J Exp Clin Med. 2016;33(3):157-61..
Preventing the clonal dissemination of microorganisms and improvement in the healthcare quality requires strategic prevention efforts and well-known infection control practices. For example, applying hand hygiene and equipment cleaning rules and creating staff awareness are particularly important to prevent the spread of an infection1414. Dettori M, Piana A, Deriu MG, Lo Curto P, Cossu A, Musumeci R, et al. Outbreak of multidrug-resistant Acinetobacter baumannii in an intensive care unit. New Microbiol. 2014:37(2):185-91.,1515. Gokmen T, Akcimen B, Kayar B, Marzi M, Koksal F. The outbreak of Acinetobacter baumannii producing OXA-23 and OXA-51 type carbapenemases in a state hospital. J Exp Clin Med. 2016;33(3):157-61..
The results of this study demonstrate that carbapenem resistance was common to all A. baumannii isolates studied, and that all isolates had MDR bacteria. The genes bla OXA-23 and bla OXA-51 were detected in all carbapenemase-producing isolates, and all these isolates belonged to the same clone. Of all the antimicrobials tested, polymyxin B and tigecycline were the most effective antimicrobials for MDR-AB. The limitations identified in our study were the relatively small number of isolates from patients, who had several comorbidities and some of the patients were hospitalized multiple times; and the limited quality of the clinical data, which was insufficient to meet the study aims.
Bacterial resistance is an emerging problem requiring the utmost attention and effort towards its mitigation. Our study highlights the need for screening colonized or infected patients and for providing frequent training to healthcare professionals in both ICUs and clinics. Moreover, surveillance for imipenem- and meropenem-resistant A. baumannii and rational administration of antimicrobials should be reinforced.
REFERENCES
-
1Founou RC, Founou, LL, Essack SY. Clinical and economic impact of antibiotic resistance in developing countries: A systematic review and meta-analysis. PLoS One. 2017;12(12):1-18.
-
2Chhatwal KWP, Vonbe RP. Nosocomial outbreaks caused by Acinetobacter baumannii and Pseudomonas aeruginosa: Results of a systematic review. Am J Infect Control. 2018;46(6):643-8.
-
3Sutherland N, Barber S. O'Neill. Review into Antibiotic Resistance. House of Commons Library, F1, Issue 1. 2017. Available from: https://research briefings .parliament.uk/ Research-Briefing/Summary/CDP-2017-0074
» https://research briefings .parliament.uk/ Research-Briefing/Summary/CDP-2017-0074 -
4World Health Organization (WHO). Chemotherapy of leprosy for control programmes. Technical Report Series 675. Geneva: WHO; 1982. 36 p.
-
5Castilho SRA, Godoy CSM, Guilarde AO, Cardoso JL, André MCP, Junqueira KAP, et al. Acinetobacter baumannii strains isolated from patients in intensive care units in Goiânia, Brazil: Molecular and drug susceptibility profiles. PLoS One . 2017;12(5):1-13.
-
6Gales AC, Castanheira M, Jones RN, Sader HS. Antimicrobial resistance among Gram-negative bacilli isolated from Latin America: results from SENTRY Antimicrobial Surveillance Program (Latin America, 2008-2010). Diagn Microbiol Infect Dis. 2012;73(4):354-60.
-
7Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol. 2018;16(2):91-102.
-
8Brazilian Committee on Antimicrobial Susceptibility Testing (BrCAST). Tabelas de pontos de corte para interpretação de CIMs e diâmetros de halos. Versão. 2020;8:2.
-
9Brasiliense Danielle, Cayô Rodrigo, Streling Ana Paula, Nodari Carolina S, Barata Rafael R, Lemos Poliana S, et al. Diversity of metallo-β-lactamase-encoding genes found in distinct species of Acinetobacter isolated from the Brazilian Amazon Region. Mem Inst Oswaldo Cruz [Internet]. 2019;114(1):1-6.
-
10Dice LR. Measures of the Amount of Ecologic Association Between Species. Ecology. 1945;26(3):297-302.
-
11Genteluci GL, Gomes DBC, Souza MJ, Carvalho KR, Villas BMHS. Emergence of polymyxin B-resistant Acinetobacter baumannii in hospitals in Rio de Janeiro. J Bras Patol Med Lab. 2016;52(2):91-5.
-
12Shin JA, Chang YS, Kim HJ, Kim SK, Chang J, Ahn CM, Byun MK. Clinical Outcomes of Tigecycline in the Treatment of Multidrug-Resistant Acinetobacter baumannii Infection. Yonsei Med J. 2012;53(5):974-84.
-
13Schuertz KF, Tuon FF, Palmeiro JK, Conte D, Telles JPM, Trevisoli LE, et al. Bacteremia and meningitis caused by OXA-23-producing Acinetobacter baumannii - molecular characterization and susceptibility testing for alternative antibiotics. Braz J Microbiol. 2018;49(s1):199-204.
-
14Dettori M, Piana A, Deriu MG, Lo Curto P, Cossu A, Musumeci R, et al. Outbreak of multidrug-resistant Acinetobacter baumannii in an intensive care unit. New Microbiol. 2014:37(2):185-91.
-
15Gokmen T, Akcimen B, Kayar B, Marzi M, Koksal F. The outbreak of Acinetobacter baumannii producing OXA-23 and OXA-51 type carbapenemases in a state hospital. J Exp Clin Med. 2016;33(3):157-61.
-
Financial Support: This research was self-funded.
Publication Dates
-
Publication in this collection
13 Nov 2020 -
Date of issue
2021
History
-
Received
22 Mar 2020 -
Accepted
18 June 2020