Abstract
In order to study the infectious agents causing human disseminated cryptococcosis in the state of Pará, North Brazil, 56 isolates of Cryptococcusspp. (54 isolated from cerebral spinal fluid and two from blood cultures) from 43 cases diagnosed between 2003-2007 were analysed. The species were determined through morphological and physiological tests and genotypes were determined by URA5-RFLP and PCR-fingerprinting (wild-type phage M13). The following species and genotypes were identified: Cryptococcus neoformans VNI (28/56, 50%), Cryptococcus gattii VGII (25/56, 44.64%) and C. gattii VGI (3/56, 5.26%). The genotype VNI occurred in 12 out of 14 HIV-positive adults, whereas the genotype VGII occurred in 11 out of 21 HIV-negative adults (p < 0.02, OR = 6.6 IC95% 0.98-56.0). All patients less than 12 years old were HIV negative and six cases were caused by the VGII genotype, one by the VGI and one by VNI. Therefore, endemic primary mycosis in HIV-negative individuals, including an unexpectedly high number of children, caused by the VGII genotype deserves further study and suggests the need for surveillance on cryptococcal infection in the state of Pará, Eastern Amazon.
Cryptococcus gattii; VGII genotype; endemic mycosis; children; state of Pará
ARTICLES
Primary endemic Cryptococcosis gattii by molecular type VGII in the state of Pará, Brazil
Wallace Raimundo A dos SantosI, II; Wieland MeyerVI; Bodo WankeV; Solange PS Evangelista CostaII; Luciana TrillesV; Jose Luiz M NascimentoII; Rita MedeirosIII; Bernardina Pernarrieta MoralesV; Claudia de Carvalho Falci BezerraV; Regina Célia Lima de MacêdoV; Silvana O FerreiraIII; Gláucia Gonçalves BarbosaV; Mauricio A PerezVII; Marília Martins NishikawaIV; Márcia dos Santos LazéraV, + + Corresponding author: marcia.lazera@ipec.fiocruz.br
IDepartamento de Farmácia, Universidade Federal do Pará, Belém, PA, Brasil
IICentro de Ciências Biológicas, Universidade Federal do Pará, Belém, PA, Brasil
IIIHospital Universitário João de Barros Barreto, Universidade Federal do Pará, Belém, PA, Brasil
IVInstituto Nacional de Controle de Qualidade em Saúde
VLaboratório de Micologia do Instituto de Pesquisa Evandro Chagas-Fiocruz, Av. Brasil 4365, 21045-900 Rio de Janeiro, RJ, Brasil
VIUniversity of Sydney Western Clinical School, Westmead Millenium Institute, Westmead Hospital, Sydney, Austrália
VIICentro de Estudos da Saúde Comunitária, Universidade Federal do Rio de Janeiro, Rio de Janeiro, RJ, Brasil
ABSTRACT
In order to study the infectious agents causing human disseminated cryptococcosis in the state of Pará, North Brazil, 56 isolates of Cryptococcusspp. (54 isolated from cerebral spinal fluid and two from blood cultures) from 43 cases diagnosed between 2003-2007 were analysed. The species were determined through morphological and physiological tests and genotypes were determined by URA5-RFLP and PCR-fingerprinting (wild-type phage M13). The following species and genotypes were identified: Cryptococcus neoformans VNI (28/56, 50%), Cryptococcus gattii VGII (25/56, 44.64%) and C. gattii VGI (3/56, 5.26%). The genotype VNI occurred in 12 out of 14 HIV-positive adults, whereas the genotype VGII occurred in 11 out of 21 HIV-negative adults (p < 0.02, OR = 6.6 IC95% 0.98-56.0). All patients less than 12 years old were HIV negative and six cases were caused by the VGII genotype, one by the VGI and one by VNI. Therefore, endemic primary mycosis in HIV-negative individuals, including an unexpectedly high number of children, caused by the VGII genotype deserves further study and suggests the need for surveillance on cryptococcal infection in the state of Pará, Eastern Amazon.
Key words:Cryptococcus gattii - VGII genotype - endemic mycosis - children - state of Pará
Cryptococcosis is a significant life-threatening fungal infection that affects humans and a large variety of animals. It is caused by two species of Cryptococcus, C. neoformans (serotypes A, D and the hybrid AD) and C. gattii (serotypes B and C). C. gattii, previously regarded as a variety of C. neoformans, is now recognized as a distinct species (Kwon-Chung & Varma 2006).
C. neoformans-induced cryptococcosis occurs worldwide and is an important cause of morbidity and mortality in immunocompromised individuals, especially AIDS patients. Cryptococcosis caused by C. gattii, however, is mainly seen in non-immunocompromised patients (La-zera et al. 2005) and regarding its geographical distribution, it is considered a tropical pathogen. However, the recent description of the Vancouver Island (Canada) outbreak suggests that C. gattii has adapted to more temperate environments (Kidd et al. 2004).
The infections caused by C. neoformans and C. gattii, as well as their corresponding serotypes and molecular types, present with different clinical symptoms and have a diverse prognosis. Therefore, an understanding of the prevailing species and types in specific geographic regions is important. The mycosis caused by C. neoformans serotype A is widespread and predominantly infects immunocompromised individuals. However, C. neoformans serotype D is predominantly found in Europe and mainly infects elderly subjects, although it has been rarely detected in Brazil (Dromer et al. 1996, Nishikawa et al. 2003).
Cryptococcosis caused by C. gattii (mainly serotype B) has been predominantly detected in immunocompetent patients and presents as a regressive lung lesion which is usually undetected, or as a peripheral lung nodule which has to be differentiated from a malignant tumour. Both species cause a basal meningoencephalitis, with or without lung lesions, that usually causes an increased intracranial pressure, cranial nerve damage and hydrocephalus. In HIV-negative patients the infection may also cause blindness and hearing loss. Cryptococcosis caused by C. gattii typically evolves sub-acutely or chronically and is often confused with viral or bacterial meningoencephalitis or other infections, including tuberculosis (Lazera et al. 2005, Lin & Heitman 2006).
C. neoformans serotype A is ubiquitous and has been associated with organic material in pigeon habitats, captive birds in domestic environments, domestic dust and wood decay in hollow trees of different species (Passoni et al. 1998, Nishikawa et al. 2003). In Brazil, it is the most prevalent serotype in clinical (89%) and environmental isolates (90.2%) in the South, Southeast and Central West regions, whereas C. gattii predominates in the Northeast region in clinical and environmental isolates (64%) (Nishikawa et al. 2003).
Most clinical isolates are haploid and heterothallic but some may be aneuploid or even diploid. The mating type of Cryptococcus is determined by a one-locus, two allele system (MAT ± and MAT a) that produces viable progeny in vitro in a 1:1 ratio. Clinical and environmental isolates are predominantly MAT ±, which has been suggested to be of higher virulence in animal models (Kwon-Chung & Bennett 1992).
PCR fingerprinting is being used in ongoing global surveys to study the epidemiology of cryptococcosis. By using primers for the minisatellite-specific core sequence of the wild-type phage M13, in combination with URA5-RFLP analysis, eight major molecular types (VN I-IV for C. neoformans and VG I-IV for C. gattii) have been identified (Meyer et al. 2003). The corresponding genotypes of these molecular types were identified using amplified fragment length polymorphism PCR according to Boekhout et al. (2001).
In the Southeast and South regions of Brazil, where consistent data are available, human cryptococcosis is caused predominantly by C. neoformans serotype A, VNI and is associated with AIDS (Casali et al. 2003, Igreja et al. 2004). However, the few studies performed in the Northeast region suggest epidemiological differen-ces, particularly the occurrence of meningitis by C. gattii serotype B in native adults and children in this region (Lazera et al. 2005). Additionally, one study performed in the North region, in Belém do Pará, also describes cryptococcosis in immunocompetent children (Corrêa et al. 1999). To increase the understanding of cryptococcosis in the North region of Brazil, we performed a retrospective and prospective study of isolates obtained from patients with meningitis, identifying the species, serotypes and molecular types of Cryptococcus. Clinical and epidemiological data were also analysed providing an epidemiological view on cryptococcosis and its causative agents in the Eastern Amazon.
MATERIALS AND METHODS
Cryptococcus identification - From May 2003-April 2007, isolates of Cryptococcus spp. isolated from patients admitted to the João Barros Barreto Hospital, a reference centre in the city of Belém, were identified by morphological and physiological tests, including phenol oxidase production on niger seed agar (NSA), cyclohe-ximide sensitivity, assimilation of carbon and nitrogen sources (Vitek ICB, bioMerieux, Durham, USA) and growth in canavanine-glycine-bromothymol blue (CGB) media. Additionally, the Crypto-Chek Kit (Iatron Laboratories, Tokyo, Japan) was used to discriminate between different serotypes of the same species.
Statistical analysis - The clinical and fungal data from the isolates were analysed with the chi-square test (Yates correction). All p-values less than 0.05 were considered significant.
Reference strains - The following set of standard laboratory strains, representing each molecular type, were used as controls in PCR-fingerprinting and URA5-RFLP: WM 148 (serotype A, VNI), WM 626 (serotype A, VNII), WM 628 (serotype AD, VNIII), WM 629 (serotype D, VNIV), WM 179 (serotype B, VGI), WM 178 (serotype B, VGII), WM 161 (serotype B, VGIII) and WM 779 (serotype C, VGIV).
DNA extraction - High molecular weight DNA was extracted according to Ferrer et al. (2001). Half an inoculation loop of culture was frozen at -20°C for 1 h and then incubated at 65°C for 1 h in 0.5 mL of extraction buffer (50 mM Tris-HCl, 50 mM EDTA, 3% sodium dodecyl sulfate, 1% 2-mercaptoethanol). The lysate was extracted with phenol-chloroform-isoamyl alcohol (25:24:1, v:v:v). DNA was recovered by isopropanol precipitation, washed with 70% (v:v) ethanol and diluted in sterile water.
URA5-RFLP - PCR of the URA5 gene was conducted in a final volume of 50 ¼L. Each reaction contained 50 ng of DNA, 1X PCR buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.1 mM EDTA, 2 mM DCT, 50% glycerol, 0.1% Tween-20 ), 0.2 mM of each dATP, dCTP, dGTP and dTTP (Roche Diagnostics GmbH), 2 mM magnesium chloride, 2.5 U Taq DNA polymerase (Bioline) and 25 ng of each primer URA5 (5' ATGTCCTCCCAAGCCCTCGACTCCG 3') and SJ01 (5' TTAAGACCTCTGAACACCGTACTC 3'). PCR was performed for 35 cycles in a Perkin-Elmer thermal cycler (model 480) using the following parameters; 94°C for 5 min initial denaturation, 35 cycles denaturation at 94°C for 45 s, 1 min annealing at 63°C and 2 min extension at 72°C, followed by a final extension cycle for 10 min at 72°C. PCR products were double digested with Sau96I (10 U/µL) and HhaI (20 U/µl) for 3 h and the fragments were separated on a 3% agarose gel stained with ethidium bromide. RFLP patterns were assigned visually by comparison with the patterns obtained from the reference strains described above.
PCR-fingerprinting - PCR-fingerprinting reactions were conducted in a volume of 50 µL containing 25 ng genomic DNA, 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl, 0.2 mM each of dATP, dCTP, dGTP and dTTP (Roche Diagnostics GmbH, Mannheim, Germany), 3 mM magnesium acetate, 30 ng primer (5' GAGGGTGGCGGTTCT 3'), and 2.5 U Amplitaq DNA polymerase (Applied Biosystems, Foster City, CA). PCR was performed for 35 cycles in a Perkin-Elmer thermal cycler (model 480) with 20 s of denaturation at 94°C, 1 min annealing at 50°C and 20 s extension at 72°C, followed by a final extension cycle for 6 min at 72°C. Amplification products were concentrated to approximately 15 µL and separated by electrophoresis on 1.4% agarose gels stained with ethidium bromide (Meyer et al. 2003). PCR-fingerprinting profiles were visually compared to the reference strains to determine the molecular types. The genetic relationships of the isolates were analysed using the 1D gel analysis module (BioGalaxy [BioAware, Hannut, Belgium]) in BioloMICS version 7.5.30 (BioAware). Similarity coefficients were calculated by using the Dice algorithm and cluster analyses were performed with the Unweighted Pair Group Method with Arithmetic mean.
Molecular determination of the mating type - The MAT ± and MAT a pheromones were determined in all isolates by PCR according to Chaturvedi et al. (2000), using the ±-mating type-specific primers 5' - CTTCACTGCCATCTTCACCA-3' and 5' -GACACAAAGGGTCATGCCA-3', and the a-mating type-specific primers 5' - CGC CTT CAC TGC TAC CTT CT-3' and 5' -AAC GCA AGA GTA AGT CGG GC-3'. C. gattii isolate mating types were determined using the primers MFA2U (5' ACACCGCCTGTTACAATGGA 3') and MFA2L (5' CAGCGTTTGAAGATGGACTTT 3'), as described by Fraser et al. (2003).
RESULTS
A total of 56 cryptococcal isolates from 43 patients with meningitis were analysed. The majority of these isolates (54 of 56) were isolated from the cerebral spinal fluid of patients, while only two were isolated from blood cultures. A single isolate was obtained from 31 patients; two isolates were obtained from 11 patients and three from one patient. In this cohort, 26 (60.5%) patients were male and 17 (39.5%) female. Eight (18.6%) children (< 12 years old) were all HIV negative and the remaining 35 (81.4%) patients comprised 21 (60%) HIV-negative and 14 (40%) HIV positive patients.
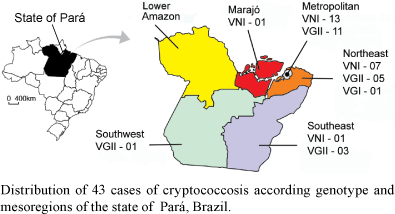
C. gattii was more frequently isolated from HIV-negative patients (19/29; 65.5%) and C. neoformans was predominantly isolated from the HIV-positive patients (12/14, 85.7%; p < 0.005; OR = 11.4; IC95% 1.8-92.2). The remaining two HIV-positive patients had C. gattii-induced cryptococcosis (Table). The identification of the species of Cryptococcus by the CGB test demonstrated that of the 56 isolates, 27 (48.2%) were C. neoformans, 20 (35.7%) were C. gattii and nine produced ambiguous results [16.1 % (Table)]. Based on the results of the serotyping analysis, 21 isolates were serotype B, 20 were serotype A and 15 were indeterminate (7 C. gattii and 8 C. neoformans). Five isolates obtained from two children from the Northeast nesoregion of Pará produced indeterminate results in both the serotyping and CGB tests. Three molecular types were identified by URA5-RFLP and PCR-fingerprinting: VNI (28/56, 50%), VGII (25/56, 44.6%) and VGI [3/56, 5.4% (Table)]. The VNI genotype was isolated from 12 of 14 HIV-positive adults, whereas the VGII genotype was isolated from 11 of 21 HIV-negative adults (p < 0.02, OR = 6.6 IC95% 0.98-56.0). All isolates were identified as MAT ± using mating type-specific primers (Fraser et al. 2003, Holliday et al. 2003). The geographical origin of the 43 cases based on molecular type and mesoregion of the state of Pará is shown in Figure.
DISCUSSION
The North region of Brazil encompasses a large area of the Amazon rainforest and more studies on human cryptococcosis and the molecular profiles of the infecting strain(s) need to be performed. A study on cryptococcosis in children in the state of Pará described 19 cases, from which nine isolates were determined to be C. gattii (Corrêa et al. 1999). In the present study, the incidence of C. gattii-induced meningitis is similar to the previously described cases in the Northeast region (Lazera et al. 2005). However, it is distinct from the cases described in the South and Southeast regions of Brazil (Nishikawa et al. 2003).
In the state of Pará, C. gattii was the main causative agent of meningitis in the HIV-negative patients and occurred also in two HIV-positive patients. The high frequency of cryptococcal meningitis in HIV-negative children (8/43; 18.6%) observed from 2003-2007 supports previous studies by Corrêa et al. (1999) and suggests that cryptococcal infection occurs early in life in this region. Previous studies have demonstrated that infantile cryptococcosis accounts for a significant fraction of cryptococcosis cases in the neighbouring state of Amazonas (33%, n = 75) (Santos 2000) and the Piauí and Maranhão, states of the Northeast region (21%, n = 257) (Martins 2003). Since the present study is based on isolates obtained from patients representing only part of the cases, the real occurrence of this mycosis is largely underestimated.
We were unable to identify the Cryptococcus species in a significant number of isolates using the CGB test. Interfering factors, such as contamination or excessive inoculum, were excluded by re-plating on NSA medium and carefully repeating the test, which yielded the same results. Similarly, a significant number of isolates were untypable by serotyping. These results demonstrate the occurrence of atypical phenotypes among clinical isolates in the state of Pará, which may cause difficulties in their species identification at the local public laboratories. Usually most Cryptococcus isolates are haploid, nevertheless some diploid or aneuploid isolates may lead to unstable phenotypes, which may be recombinants or hybrids (Kwon-Chung & Varma 2006). Further genetic studies are necessary to evaluate the atypical clinical isolates from Pará.
The molecular type VGII was found to be the main causative agent in cases of C. gattii-induced cryptococcosis. These findings are in agreement with a recent Brazilian study demonstrating that the VGII genotype is the main cause of endemic cryptococcosis in immunocompetent hosts in different states of the North and Northeast regions of Brazil (Trilles et al. 2008).
Most cases caused by the VGII and VNI types were from the metropolitan and Northeast mesoregions of Pará, both near the Hospital Universitário João de Barros Barreto. VGII and VNI types were also identified in more distant mesoregions of Pará such as Southeast, Southwest and Marajó. However, in the Lower Amazon mesoregion no case was identified, which may be due to the large distance that must be travelled (> 12 h by boat) to reach health services or the reference centre in the city of Belém. For the first time we demonstrate the occurrence and predominance of the molecular type VGII as an agent for cryptococcosis in normal individuals living in the state of Pará, including a significant number of paediatric cases. These paediatric cases came from different mesoregions, suggesting that the environmental sources of VGII are widespread across the state. Additionally, one VGI case in a child from the Northeast mesoregion indicates that this molecular type may be also present in the local environment.
The mesoregions of Pará, with the exception of the metropolitan mesoregion, are mainly occupied by small communities and villages in the Amazon rainforest and share areas for cattle pastures and wood exploitation. The metropolitan mesoregion contains the areas of urbanization surrounding the city and harbour of Belém. At this port of the Amazon estuary, an extensive trade of wood and wood by-products, both for local use and for export, takes place. In Brazil, C. gattii and C. neoformans have been described in the decaying heartwood of a number of tree species, a possible primary ecological niche for both pathogens (Lazera et al. 2000). C. gattii was isolated from wood decay in a native jungle tree (Guettarda acreana) in Maracá, a river island without human activity in the Occidental Amazon rainforest, North Brazil (Fortes et al. 2001). This finding provides evidence that wild tropical forests may harbour C. gattii. The C. gattii isolated from the Guettarda tree was further identified as genotype AFLP6 (Trilles et al. 2003), which corresponds to the molecular type VGII.
C. gattii AFLP6/VGII seems to be well-adapted to the semiarid region in Northeastern Brazil, where it was isolated from the spleen of one armadillo, from wood decay in different trees and from human clinical specimens (Trilles et al. 2003). The genotype VGII is also the causative agent of the largest known outbreak infecting humans and animals and the first reported in a temperate area, Vancouver Island, Canada (Kidd et al. 2004). Considering the adaptive potential, expansive behaviour and pathogenicity of this molecular type, it is necessary to place accessible medical units in areas far from the city of Belém. These units will be able to provide specific medical information and laboratories for the early diagnosis, fungal isolation, identification and storage of the isolates. Additionally, these units can function as a registration system for the active surveillance of cryptococcosis in the state of Pará, Brazil.
ACKNOWLEDGEMENTS
To Manuela da Silva, for revising the manuscript.
Received 5 August 2008
Accepted 13 November 2008
Financial support: NH&MRC (grant 352303), Canberra, Australia to WM, FIOCRUZ, CAPES, PQI program to FIOCRUZ/UFPA/NMT, CNPq (grant 478990/2006-5).
- Boekhout T, Theelen B, Diaz M, Fell JW, Hop WCJ, Abeln ECA, Dromer F, Meyer W 2001. Hybrids genotypes in the pathogenic yeast Cryptococcus neoformans Microbiology 147: 891-907.
- Casali AK, Goulart L, Rosa e Silva LK, Ribeiro AM, Amaral AA, Alves SH, Schrank A, Meyer W, Vainstein H 2003. Molecular typing of clinical and environmental Cryptococcus neoformans isolates in the Brazilian state of Rio Grande do Sul. FEMS Yeast research 3: 405-415.
- Chaturvedi S, Rodeghier D, Fan J, Mc Vleilland CM, Wickes BL, Chaturvedi V 2000. Direct PCR of Cryptococcus neoformans Mat± and Mat a pheromones to determine mating type, ploidy and variety: a tool for epidemiological and molecular pathogenesis studies. J Clin Microbiol 38: 2007-2009.
- Correa MP, Oliveira EC, Duarte RR, Pardal PP, Oliveira FM, Severo LC 1999. Cryptococcosis in children in the state of Pará, Brazil. Rev Soc Bras Med Trop 32: 505-508.
- Dromer F, Mathoulin S, Dupont B, Laporte A, the French Crytococcosis Study Group 1996. Epidemiology of cryptococcosis in France: a 9-year survey (1985-1993). Clin Infect Dis 23: 82-90.
- Ferrer C, Colom F, Frase's S, Mulet E, Abad L, Alio JL 2001. Detection and identification of fungal pathogens by PCR and by ITS2 and 5.8S ribosomal DNA typing in ocular infections. J Clin Microbiol 39: 2873-2879.
- Fortes ST, Lazera MS, Nishikawa MM, Macedo RCL, Bodo Wanke 2001. First isolation of Cryptococcus neoformans var. gattii from native jungle tree in the Brazilian Amazon rainforest. Mycoses 44: 137-140.
- Fraser JA, Subaran RL, Nichols CB, Heitman J 2003. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot Cell 2: 1036-1045.
- Halliday CL, Bui T, Krocenberger M, Malik R, Ellis DH, Carter D 2003. Clonal reproduction and limited dispersal in an environmental population of Cryptococcus neoformans var. gattii isolates from Australia. J Clin Microbiol 41: 703-711.
- Igreja RP, Lazera MS, Wanke B, Galhardo MC, Kidd SE, Meyer W 2004. Molecular epidemiology of Cryptococcus neoformans isolates from AIDS patients of the Brazilian city, Rio de Janeiro. Med Mycol 42: 229-238.
- Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, MacDougall L, Boekhout T, Kwon-Chung KJ, Meyer W 2004. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). PNAS 101: 17258-17263.
- Kwon-Chung KJ, Bennett JE 1992. Cryptococcosis. In Kwon- Chung KJ & Bennett JE (eds.), Medical Mycology, 1st ed., Lea & Febiger, Philadelphia, p. 392-446.
- Kwon-Chung KJ, Varma SA 2006. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res 6: 574-587.
- Lazera MS, Cavalcanti MAS, Londero AT, Trilles L, Nishikawa MM, Wanke B 2000. Possible primary niche of Cryptococcus neoformans Med Micol 38: 379-383.
- Lazera MS, Gutierrez-Galhardo MC, Cavalcanti MAS, Wanke B 2005. Criptococose. In JR Coura (org), Dinâmica das Doenças Infecciosas e Parasitárias, Vol. II, 1st ed., Guanabara Koogam, Rio de Janeiro, p. 1223-1235.
- Lin X, Heitman J 2006. The Biology of the Cryptococcus neoformans Species Complex. Ann Rev Microbiol 60: 69-105.
- Martins LMS 2003. Epidemiologia da criptococose em crianças e adultos jovens e diversidade de Cryptococcus neoformans no Meio Norte do Brasil, MSc Thesis, Instituto Oswaldo Cruz-Fiocruz, Rio de Janeiro, 87 pp.
- Meyer W, Castañeda A, Huynh JS, Castañeda E, IberoAmerican Cryptococcal Study Group 2003. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg Infect Dis 9: 189-195.
- Nishikawa MM, Lazera MS, Barbosa GG, Trilles L, Balassiano BR, Macedo RCL, Bezerra CF, Perez MA, Cardarelli P, Wanke B 2003. Serotyping of 467 Cryptococcus neoformans isolates from clinical and environmental sources in Brazil: Analysis of host and regional patterns. J Clin Microbiol 41: 73-77.
- Passoni LFC, Wanke B, Nishikawa MM, Lazera MS 1998. Cryptococcus neoformans isolated from human dwellings in Rio de Janeiro, Brazil: an analysis of the domestic environment of AIDS patients with and without cryptococcosis. Med Mycol 36: 305-311.
- Santos LO 2000. Criptococose no estado do Amazonas: estudo de 75 casos diagnosticados na Fundação de Medicina Tropical/FMT/IMTM, Manaus, AM (1988-1998), Msc Thesis, Instituto Oswaldo Cruz-Fiocruz, Rio de Janeiro 154 pp.
- Trilles L, Lazera M, Wanke B, Oliveira RV, Barbosa GG, Nishikawa MM, Morales BP, Meyer W 2008. Regional pattern of the molecular types of Cryptococcus neoformans and Cryptococcus gattii in Brazil. Mem Inst Oswaldo Cruz 103: 455-462.
- Trilles L, Lazera M, Wanke B, Theelen B, Boekhout T 2003. Genetic characterization of environmental isolates of the Cryptococcus neoformans species complex from Brazil. Med Mycol 41: 383-390.
Publication Dates
-
Publication in this collection
13 Jan 2009 -
Date of issue
Dec 2008
History
-
Received
05 Aug 2008 -
Accepted
13 Nov 2008