Abstract
The aim of this work was to establish a modified pre-diagnostic polymerase chain reaction (PCR) protocol using a single primer set that enables successful amplification of a highly conserved mammalian sequence in order to determine overall sample DNA quality for multiple mammalian species that inhabit areas endemic for leishmaniasis. The gene encoding interphotoreceptor retinoid-binding protein (IRBP), but not other conserved genes, was efficiently amplified in DNA samples from tail skin, ear skin, bone marrow, liver and spleen from all of the species tested. In tissue samples that were PCR-positive for Leishmania, we found that DNA from 100%, 55% and 22% of the samples tested resulted in a positive PCR reaction for the IRBP, beta-actin and beta-globin genes, respectively. Nucleotide sequencing of an IRBP amplicon resolved any questions regarding the taxonomical classification of a rodent, which was previously based simply on the morphological features of the animal. Therefore, PCR amplification and analysis of the IRBP amplicon are suitable for pre-diagnostically assessing DNA quality and identifying mammalian species living in areas endemic to leishmaniasis and other diseases.
Leishmania spp; PCR; diagnostic; IRBP
ARTICLES
IInstituto de Pesquisas René Rachou-Fiocruz, Av. Augusto de Lima 1715, 30190-002 Belo Horizonte, MG, Brasil
IIWorld Health Organization Collaborating Centre for Leishmaniasis, Servicio de Parasitología, Centro Nacional de Microbiología, Instituto de Salud Carlos III, Madrid, Spain
IIILaboratório de Leishmanioses, Departamento de Parasitologia
IVLaboratório de Genes Inflamatórios, Departamento de Morfologia, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Belo Horizonte, MG, Brasil
ABSTRACT
The aim of this work was to establish a modified pre-diagnostic polymerase chain reaction (PCR) protocol using a single primer set that enables successful amplification of a highly conserved mammalian sequence in order to determine overall sample DNA quality for multiple mammalian species that inhabit areas endemic for leishmaniasis. The gene encoding interphotoreceptor retinoid-binding protein (IRBP), but not other conserved genes, was efficiently amplified in DNA samples from tail skin, ear skin, bone marrow, liver and spleen from all of the species tested. In tissue samples that were PCR-positive for Leishmania, we found that DNA from 100%, 55% and 22% of the samples tested resulted in a positive PCR reaction for the IRBP, beta-actin and beta-globin genes, respectively. Nucleotide sequencing of an IRBP amplicon resolved any questions regarding the taxonomical classification of a rodent, which was previously based simply on the morphological features of the animal. Therefore, PCR amplification and analysis of the IRBP amplicon are suitable for pre-diagnostically assessing DNA quality and identifying mammalian species living in areas endemic to leishmaniasis and other diseases.
Key words:Leishmania spp - PCR - diagnostic - IRBP
Most techniques that are used in molecular analysis require DNA of good quality, which is purified from polymerase chain reaction (PCR) inhibitors that can be carried over into the reaction (Greer et al. 1991). More importantly, an internal control gene of host origin must be amplified in a pre-diagnostic PCR reaction to determine the overall DNA quality of the sample; this is of particular value in epidemiological pathogen surveys for veterinary medical applications. Ideally, the selected gene would have high sequence conservation among several mammalian species of interest to the study, allowing for the use of one host control gene and therefore a single primer set for the pre-diagnostic PCR assessment of DNA quality in all samples. Diagnostic advancements on this front are often hindered by incomplete sequence data or sequence annotation for a number of mammalian host species of interest.
In this paper, we discuss our search for candidate control genes in eight local mammalian hosts of the Leishmania parasite. Oligonucleotide sequences specific for highly conserved genes, such as beta-actin and beta-globin, have previously been used to detect DNA from a large number of mammalian species (du Breuil et al. 1993, Tohgi et al. 1998). However, despite high-throughput genomic DNA sequencing of several organisms, there is still inadequate nucleotide sequence information available for these and other mammals. This information is required to design oligonucleotides that anneal to the most conserved regions of DNA. Therefore, we sought to establish an alternative PCR protocol in which a unique pair of primers could be used to amplify DNA from different tissues of a number of mammalian species, including dogs, rodents and marsupials that are important in the epidemiology of leishmaniasis.
MATERIALS AND METHODS
Four highly conserved genes, cytochrome b, histone H4, 18S RNA and interphotoreceptor retinoid-binding protein (IRBP), were chosen in addition to beta-actin and beta-globin genes and Genbank was searched for gene sequences of the mammalian species in our study. The gene sequences for cytochrome b and IRBP were the only sequences found in the database for all of the species included in our study. However, IRBP nucleotide sequences were more similar among species as compared to other gene sequences.
Following alignment of a number of IRBP gene sequences, we found that the most conserved regions were within exon 1 and we designed primers based on the canine sequence due to its relevance to veterinary medical applications. As shown in Figure, there is a high degree of similarity among the nucleotide sequences in the region used for primer annealing.
Samples of tissues (ear skin, tail skin, liver, spleen and bone marrow) from rodents (Mus musculus, Rattus rattus, Rattus novergicus, Necromys lasiurus and Cerradomys subflavus), marsupials (Didelphis albiventris) and domestic dogs and cats were evaluated for infection with different species of Leishmania. The animals were captured in an area endemic for leishmaniasis in Belo Horizonte, Minas Gerais, Brazil and samples were collected in accordance with the ethical principles for animal experimentation of the Brazilian College of Animal Experimentation. Clinical specimens from dogs were obtained as previously described (Quaresma et al. 2009).
We have established that storing tissue samples in 70% ethanol at -20ºC rather than in 100% is suitable for tissue preservation until DNA extraction, which was performed using the Cells and Tissue Prep Genomic DNA Isolation kit® (GE Healthcare, Little Chalfont, Buckinghamshire, UK), according to the manufacturer's instructions. The Column Chromatography-GFX Genomic DNA Blood Purification System (GE Healthcare, Little Chalfont, Buckinghamshire, UK) was used to obtain DNA from the bone marrow, according to the manufacturer's instructions. DNA extracted from clinical samples was subjected to PCR with primers targeting the 120-bp conserved region of the Leishmania kDNA minicircle as described in Degrave et al. (1994) and Quaresma et al. (2009).
We designed primers for IRBP based on highly conserved stretches within exon 1 of the canine gene sequence to amplify a 227-bp fragment (nucleotide positions 400-626, GeneBank accession DQ205906). The efficiency of the IRBP primers was compared with that of beta-actin and beta-globin primers. The nucleotide sequences of the primers used in this study and the PCR conditions were as follows: IRBP-CF-FWD (5'-TCCAACACCACCACTGAGATCTGGAC-3') and IRBP-CF-REV (5'-GTGAGGAAGAAATCGGACTGGCC-3') for IRBP amplification, Actin-FWD (5'-CGGAACCGCTCATTGCC-3') and Actin-REV (5'-ACCCACACTGTGCCCATCTA-3') for beta-actin amplification (du Breuil et al. 1993, Tohgi et al. 1998), Glob-FWD (5'-CAACTTCATCCACGTTCACC-3') and Glob-REV (5'-CACAACTGTGTTCACTAGC-3') for beta-globin amplification (Greer et al. 1991, Quaresma et al. 2009). The cycling conditions for IRBP and beta-actin were carried out with a first denaturation step at 95ºC for 4 min, followed by 35 cycles at 94ºC for 30 s, 57ºC for 30 s, 72ºC for 1 min and then a final extension step at 72ºC for 5 min. For beta-globin amplification, the conditions were similar, except that the annealing temperature was 40ºC and the extension time was 45 s. Two PCR buffer conditions were established for the amplification of target genes. In condition A, the reactions were at a final volume of 25 µL and contained 2 µL of DNA sample, 2.5 µL of 10X PCR buffer solution [670 mM Tris-HCl pH 8.8, 67 mM MgCl2, 167 mM (NH4)2SO4, 100 mM 2-mercaptoethanol], 1.5 µL of 10 mM dNTP mix, 2.5 µL of dimethyl sulfoxide (DMSO) (molecular biology grade), 1.25 µL of each primer (10 µM), 0.15 µL of Taq DNA polymerase (Amersham Biosciences, 5 U/µL) and 15.1 µL of sterile distilled H2O. In condition B, the Ready-To-Go PCR Beads® kit (GE Healthcare, Little Chalfont, Buckinghamshire, UK) was used. The final concentration of reagents in this kit, as provided by the manufacturer, were as follows: 10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, 200 µM of dNTP mix, 2.5 units of Taq DNA polymerase (Amersham Biosciences, 5 U/µL) and stabilizers, including bovine serum albumin. DMSO was also added to the reactions (2.5 µL per reaction). An aliquot of each primer (10 pmoles) was added to the PCR mix. Each reaction contained 2 µL of DNA sample in a final volume of 25 µL. PCRs were performed in the Personal Master Thermocycler (Applied Biosystems GeneAmp System 2400) using the conditions described above. PCR products were fractionated on 6% polyacrylamide gels followed by silver staining (0.2% AgNO3), using standard procedures (Quaresma et al. 2009), or on 1.5% agarose gels, followed by visualization with ethidium bromide. For IRBP amplification, we found that condition B consistently yielded better results. Conversely, condition A yielded better results for beta-actin and beta-globin amplifications.
Further experiments were carried out to optimize the PCR for IRBP. The annealing temperature (60ºC) was calculated and experimental temperatures were tested by increasing or decreasing that temperature by 3ºC-intervals. The best amplification result occurred with a temperature of 57ºC.
RESULTS
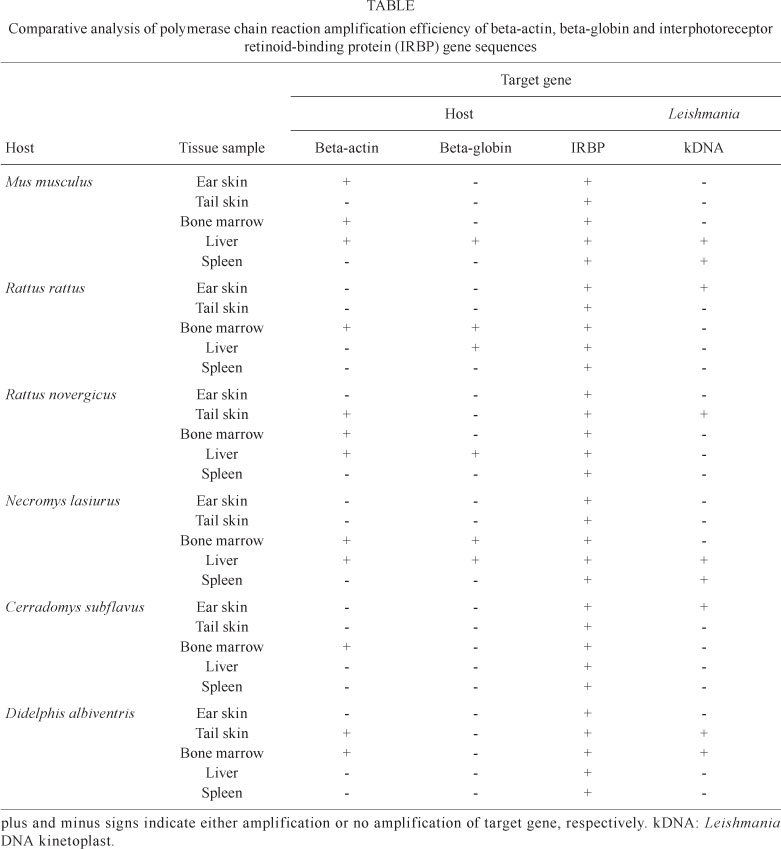
We analysed DNA from various tissue samples isolated from at least six different mammalian species that were either naturally infected or not infected with Leishmania. The results (Table) show that IRBP was efficiently amplified from all DNA samples, regardless of the tissue type or mammalian species. Furthermore, beta-actin was amplified from all bone marrow DNA samples from all mammalian species tested. However, no beta-actin specific sequence was amplified from spleen DNA samples. For liver samples, three mammalian species had positive reactions for beta-actin. For ear and tail skin samples, there were beta-actin-positive reactions for only one and two species, respectively. Beta-globin amplification was much less efficient compared to IRBP and beta-actin amplification. There was no beta-globin DNA amplification in ear skin, tail skin or spleen samples. Beta-globin was only amplified in DNA samples from the liver of four mammalian species. In DNA samples from bone marrow, only two species had beta-globin-positive reactions.
Next, we analysed those mammalian species with tissue samples that were PCR-positive for Leishmania kDNA for DNA amplification of the three target genes. We found that IRBP amplification was remarkably efficient (100%) in all nine DNA tissue samples that were PCR-positive for Leishmania. For beta-actin, five of nine (55%) samples were positive and thus only these samples were considered to be reliable. Amplification of beta-globin was much less efficient and only two samples (22%) yielded any result. Of the 21 tissue samples that were negative for Leishmania by PCR analysis, four (19%) were positive for beta-globin, seven (33%) were positive for beta-actin and all (100%) were positive for IRBP. These observations clearly demonstrate that reliable host DNA amplification is needed to avoid false negative results and thus should be included in diagnostic studies of different animal species, especially during the analysis of different tissues.
We took advantage of the IRBP amplicon specificity and performed nucleotide sequencing and Basic Local Alignment Search Tool analysis of DNA sequences that were isolated from at least one tissue sample for each mammalian species tested. Interestingly, one species was initially classified as Akodon; however, following sequencing of the IRBP amplicon and cytochrome b (data not shown), this species was actually confirmed to be N. lasiurus.
DISCUSSION
Advantages of using the PCR protocol for IRBP DNA amplification vs. other targets, such as beta-actin, beta-globin or cytochrome b, are that IRBP gene sequences are known for many vertebrates and have high sequence similarity among different mammalian species (Pepperberg et al. 1993, Harada et al. 1995, Stanhope et al. 1996, DeBry & Sagel 2001, Weksler 2003, Poux & Douzery 2004, Nickerson et al. 2006, Oliveira et al. 2010).
Analysis of the sequence variability from different species could be a useful tool for species identification, especially for species for which morphological analysis per se hampers a precise classification. As exemplified above, sequencing the IRBP amplicon from a particular DNA tissue sample helped us to definitively identify a Leishmania-infected N. lasiurus mammal, which had been previously identified as Akodon.
In summary, the IRBP primers provide an important tool to pre-assess the DNA quality in samples that are taken from mammalian species that inhabit areas endemic to leishmaniasis. Here, we have introduced IRBP gene-specific PCR amplification and proven its value as an initial step to increasing the confidence in the quality of DNA isolated from Leishmania-infected tissues. We believe that this work will greatly improve the confidence of PCR-based diagnoses of animals infected with Leishmania and other pathogens, as well as help to pre-assess the quality of DNA in tissue samples. In addition, this method can be used as an auxiliary tool in the identification of host species, or even to identify the source of a blood meal in female sand flies, such as has been done with cytochrome b in studies of tsetse flies (Steuber et al. 2005) or mosquitoes (Oshaghi et al. 2006).
ACKNOWLEDGEMENTS
To Olindo AM Filho, for suggestions.
Received 12 April 2010
Accepted 12 August 2010
Financial support: FIOCRUZ/PAPES IV, CNPq, CAPES, LeishEpiNetSA (European Community), FAPEMIG
- DeBry RW, Sagel RM 2001. Phylogeny of rodentia (Mammalia) inferred from the nuclear-encoded gene IRBP. Mol Phylogenet Evol 19: 290-301.
- Degrave W, Fernandes O, Campbell D, Bozza M, Lopes U 1994. Use of molecular probes and PCR for detection and typing of Leishmania - a mini-review. Mem Inst Oswaldo Cruz 89: 463-469.
- du Breuil RM, Patel JM, Mendelow BV 1993. Quantitation of beta-actin-specific mRNA transcripts using xeno-competitive PCR. PCR Methods Appl 3: 57-59.
- Greer CE, Peterson SL, Kiviat NB, Manos MM 1991. PCR amplification from paraffin-embedded tissues. Effects of fixative and fixation time. Am J Clin Pathol 95: 117-124.
- Harada ML, Schneider H, Schneider MP, Sampaio I, Czelusniak J, Goodman M 1995. DNA evidence on the phylogenetic systematics of New World monkeys: support for the sister-grouping of Cebus and Saimiri from two unlinked nuclear genes. Mol Phylogenet Evol 4: 331-349.
- Nickerson JM, Frey RA, Ciavatta VT, Stenkamp DL 2006. Interphotoreceptor retinoid-binding protein gene structure in tetrapods and teleost fish. Mol Vis 12: 1565-1585.
- Oliveira R, Castro D, Godinho R, Luikart G, Alves PC 2010. Species identification using a small nuclear gene fragment: application to sympatric wild carnivores from South-western Europe. Conserv Genet 11: 1023-1032.
- Oshaghi MA, Chavshin AR, Vatandoost H, Yaaghoobi F, Mohtarami F, Noorjah N 2006. Effects of post-ingestion and physical conditions on PCR amplification of host blood meal DNA in mosquitoes. Exp Parasitol 112: 232-236.
- Pepperberg DR, Okajima TL, Wiggert B, Ripps H, Crouch RK, Chader GJ 1993. Interphotoreceptor retinoid-binding protein (IRBP). Molecular biology and physiological role in the visual cycle of rhodopsin. Mol Neurobiol 7: 61-85.
- Poux C, Douzery EJ 2004. Primate phylogeny, evolutionary rate variations and divergence times: a contribution from the nuclear gene IRBP. Am J Phys Anthropol 124: 1-16.
- Quaresma PF, Murta SM, Ferreira E de C, da Rocha-Lima AC, Xavier AA, Gontijo CM 2009. Molecular diagnosis of canine visceral leishmaniasis: identification of Leishmania species by PCR-RFLP and quantification of parasite DNA by real-time PCR. Acta Trop 111: 289-294.
- Stanhope MJ, Smith MR, Waddell VG, Porter CA, Shivji MS, Goodman M 1996. Mammalian evolution and the interphotoreceptor retinoid binding protein (IRBP) gene: convincing evidence for several superordinal clades. J Mol Evol 43: 83-92.
- Steuber S, Abdel-Rady A, Clausen PH 2005. PCR-RFLP analysis: a promising technique for host species identification of blood meals from tsetse flies (Diptera: Glossinidae). Parasitol Res 97: 247-254.
- Tohgi H, Utsugisawa K, Yoshimura M, Nagane Y, Mihara M 1998. Age-related changes in nicotinic acetylcholine receptor subunits alpha4 and beta2 messenger RNA expression in postmortem human frontal cortex and hippocampus. Neurosci Lett 245: 139-142.
- Weksler M 2003. Phylogeny of Neotropical oryzomyine rodents (Muridae: Sigmodontinae) based on the nuclear IRBP exon. Mol Phylogenet Evol 29: 331-349.
Alternative PCR protocol using a single primer set for assessing DNA quality in several tissues from a large variety of mammalian species living in areas endemic for leishmaniasis
Publication Dates
-
Publication in this collection
18 Nov 2010 -
Date of issue
Nov 2010
History
-
Accepted
12 Aug 2010 -
Received
12 Apr 2010