Abstracts
The aim of the present study was to examine genetic variability in populations of An. cruzii by employing PCR-RAPD and PCR-RFLP markers. All analyses were carried out using individuals of the F1 generation of wild caught females obtained in Santa Catarina State (Florianópolis and São Francisco do Sul), Paraná State (Morretes, Paranaguá and Guaratuba) and São Paulo State (Cananéia). In the PCR-RAPD experiments, seven primers were used for comparisons within and among populations. The restriction profile of the ITS2 including a fragment of both 5.8S and 28S regions of the rDNA was obtained with the enzymes BstUI, HaeIII, TaqI, HhaI, Sau96I, HinfI, HincII and NruI. The PCR-RAPD method detected a large number of polymorphic bands. Genetic distance among populations of An. cruzii varied from 0,0214 to 0,0673, suggesting that all individuals used in the analyses belong to a single species. The number of migrants per generation (Nm) was 4.3, showing the existence of gene flow among populations. The restriction profile of the ITS2, 5.8S and 28S gene regions was similar in all An. cruzii samples, whereas the results obtained by using HhaI and NruI are indicative that the individuals analyzed have nucleotide sequences distinct from those of An. cruzii samples from Peruíbe and Juquiazinho deposited in GenBank.
Anophelinae; genetic variability; ITS2; molecular markers
O objetivo desse trabalho foi analisar a variabilidade genética em populações de An. cruzii utilizando as técnicas PCR-RAPD e PCR-RFLP. As análises foram realizadas a partir de adultos da geração F1 de fêmeas coletadas nos estados de Santa Catarina (Florianópolis e São Francisco do Sul), Paraná (Morretes, Paranaguá e Guaratuba) e São Paulo (Cananéia). Na PCR-RAPD, sete iniciadores foram utilizados para comparação dentro e entre populações. Os perfis de restrição da região ITS2 e parte do genes 5.8S e 28S foram obtidos com as enzimas BstUI, HaeIII, TaqI, HhaI, Sau96I, HinfI, HincII e NruI. Utilizando-se a técnica PCR-RAPD, elevado número de bandas polimórficas foi detectado. As distâncias genéticas entre as populações variaram de 0,0214 a 0,0673, sugerindo que as amostras representam uma única espécie. O número de migrantes por geração (Nm) foi de 4,3, indicando a existência de fluxo gênico entre as populações. Os padrões de restrição da região ITS2 foram semelhantes para todas as amostras de An. cruzii, entretanto, os resultados obtidos com HhaI e NruI indicam que os indivíduos analisados apresentam seqüências diferentes daquelas disponíveis no GenBank para as populações de Peruíbe e Juquiazinho.
Anophelinae; ITS2; marcadores moleculares; variabilidade genética
MEDICAL AND VETERINARY ENTOMOLOGY
PCR-RAPD and PCR-RFLP polymorphisms detected in Anopheles cruzii (Diptera, Culicidae)1 1 Contribution # 1626 of the Departamento de Zoologia, Universidade Federal do Paraná.
PCR-RAPD and PCR-RFLP polymorphisms detected in Anopheles cruzii (Diptera, Culicidae)
Daniéla CaladoI; Mario Antonio Navarro-SilvaI; Maria Anice Mureb SallumII
ILaboratório de Entomologia Médica e Veterinária, Departamento de Zoologia, UFPR. Centro Politécnico s/n, Caixa Postal 19020, 81531-980 Curitiba-PR, Brasil
IINúcleo de Pesquisas Taxonômica e Sistemática em Entomologia Médica, Departamento de Epidemiologia, Faculdade de Saúde Pública, Universidade de São Paulo, Avenida Dr. Arnaldo 715, 01246-904 São Paulo-SP, Brazil. danielacalado@hotmail.com; mnavarro@ufpr.br; masallum@usp.br
ABSTRACT
The aim of the present study was to examine genetic variability in populations of An. cruzii by employing PCR-RAPD and PCR-RFLP markers. All analyses were carried out using individuals of the F1 generation of wild caught females obtained in Santa Catarina State (Florianópolis and São Francisco do Sul), Paraná State (Morretes, Paranaguá and Guaratuba) and São Paulo State (Cananéia). In the PCR-RAPD experiments, seven primers were used for comparisons within and among populations. The restriction profile of the ITS2 including a fragment of both 5.8S and 28S regions of the rDNA was obtained with the enzymes BstUI, HaeIII, TaqI, HhaI, Sau96I, HinfI, HincII and NruI. The PCR-RAPD method detected a large number of polymorphic bands. Genetic distance among populations of An. cruzii varied from 0,0214 to 0,0673, suggesting that all individuals used in the analyses belong to a single species. The number of migrants per generation (Nm) was 4.3, showing the existence of gene flow among populations. The restriction profile of the ITS2, 5.8S and 28S gene regions was similar in all An. cruzii samples, whereas the results obtained by using HhaI and NruI are indicative that the individuals analyzed have nucleotide sequences distinct from those of An. cruzii samples from Peruíbe and Juquiazinho deposited in GenBank.
Keywords: Anophelinae; genetic variability; ITS2; molecular markers.
RESUMO
O objetivo desse trabalho foi analisar a variabilidade genética em populações de An. cruzii utilizando as técnicas PCR-RAPD e PCR-RFLP. As análises foram realizadas a partir de adultos da geração F1 de fêmeas coletadas nos estados de Santa Catarina (Florianópolis e São Francisco do Sul), Paraná (Morretes, Paranaguá e Guaratuba) e São Paulo (Cananéia). Na PCR-RAPD, sete iniciadores foram utilizados para comparação dentro e entre populações. Os perfis de restrição da região ITS2 e parte do genes 5.8S e 28S foram obtidos com as enzimas BstUI, HaeIII, TaqI, HhaI, Sau96I, HinfI, HincII e NruI. Utilizando-se a técnica PCR-RAPD, elevado número de bandas polimórficas foi detectado. As distâncias genéticas entre as populações variaram de 0,0214 a 0,0673, sugerindo que as amostras representam uma única espécie. O número de migrantes por geração (Nm) foi de 4,3, indicando a existência de fluxo gênico entre as populações. Os padrões de restrição da região ITS2 foram semelhantes para todas as amostras de An. cruzii, entretanto, os resultados obtidos com HhaI e NruI indicam que os indivíduos analisados apresentam seqüências diferentes daquelas disponíveis no GenBank para as populações de Peruíbe e Juquiazinho.
Palavras chave: Anophelinae; ITS2; marcadores moleculares; variabilidade genética.
There has been a noticeable increase during recent years of studies in morphology, genetic diversity and population genetics of Neotropical Anophelinae mosquitoes. However, most such research addresses subgenus Nyssorhynchus. In light of its epidemiological importance, similar data on subgenus Kerteszia are surprisingly few. This may be due in part to the difficulty of rearing these species in the laboratory and because sample sizes of collected mosquitoes are often low (Carvalho-Pinto & Lourenço-de-Oliveira 2003).
Within subgenus Kerteszia, An. cruzii is notable because it is the principal vector of Plasmodium spp. in the coastal south and southeast of Brazil. In the States of Paraná, Santa Catarina and São Paulo, An. cruzii specimens have been found infected with oocysts and sporozoites (Coutinho et al. 1943; Branquinho et al. 1997).
Polymorphisms detected in chromosome, allozyme and molecular markers from An. cruzii suggest the existence of a species complex. Ramirez & Dessen (2000a,b) recognized three banding patterns in polytene chromosomes in populations from coastal São Paulo and Santa Catarina States, and suggested the existence of three corresponding cryptic species, designated provisionally as An. cruzii A, B and C.
Furthermore, allozyme variants have been detected in An. cruzii populations that also support the existence of a species complex. Carvalho-Pinto & Lourenço-de-Oliveira (2004) analyzed the enzymatic profiles of several populations and confirmed that samples from Bahia State differ from those from São Paulo, Rio de Janeiro and Santa Catarina States, and could thus constitute either an isolated population or a distinct species. Mallafronte et al. (1997) noted differences in ITS2 (Second Internal Transcribed Spacer) sequence and restriction patterns in samples from five localities in São Paulo State.
A robust understanding of the genetic variability of a vector species is fundamental to understanding their population dynamics, their relationships with potential hosts and pathogenic agents, and for the determination of new taxonomic characters (as in the case of cryptic species). Our objective in the present study was an analysis of the genetic variability of samples from different An. cruzii populations using PCR-RAPD and PCR-RFLP of ITS2.
MATERIAL AND METHODS
Samples.Anopheles adults were captured in the Brazilian Coastal Atlantic Forest from May 2001 to November 2003. Females of Anopheles (Kerteszia) spp. were captured during crepuscular and nocturnal periods, except for samples from Paranaguá (25º30'S, 48º18'W), Paraná State, which were collected between 10:00 and 12:00 h. In Paraná State, collections were undertaken in the Paraná Agronomical Institute Experimental Station, in the municipality of Morretes (48º49'W, 25º30'S); in the municipality of Guaratuba; in Palmito State Park (25º35'S, 48º32'W) and in the Ilha do Mel Ecological Station, Paranaguá Municipality (25º30'S, 48º18'W). In Santa Catarina, samples were collected in the municipality of São Francisco do Sul (26º14'S, 48º41'W) and in Florianópolis in Desterro Park of the Federal University of Santa Catarina (27º47'S, 48º34'W adjacent to motorway SC-401). In São Paulo, mosquitoes were collected in the Municipality of Cananéia (24º53'S 47º50'W).
Wild-caught females were isolated and allowed to oviposit in order to produce an F1 generation that was used for identification. A portion of the reared samples were used for association of immature exuviae and corresponding adults, which were mounted and deposited in the entomological collection of Pe. Jesus Santiago Moure (DZUP) of the Zoology Department of the Federal University of Paraná. The remaining reared progeny (139 individuals) was stored in ethanol at 20 ºC for later use in molecular analyses.
DNA extraction and PCR-RAPD conditions. To avoid DNA contamination from parasites and/or host blood, only fasted adult F1 progeny were used in the molecular analyses. The DNA extraction from single mosquito samples was as in the protocol of Carvalho & Vieira (2001), which was modified from Cheung et al. (1993).
The amplification reaction contained 15-30 ng of DNA, 1X PCR buffer, 3 mM MgCl2, 100 mM of each dNTP, 5 pmol of each primer (Operon Technologies Inc. Alameda, CA.), 1U Taq DNA polymerase (Invitrogen) and the addition of Milli-Q sterile water to a final volume of 25 ml. Polymerase chain reaction was undertaken in a PTC-100TM (MJ Research, Inc. Watertown, MA) thermocycler, using the following cycling conditions: initial denaturing at 94ºC for three minutes followed by 41 amplification cycles (one minute denaturing at 94ºC, one minute annealing at 35ºC and extension at 72ºC for two minutes) and a final extension at 72ºC for four minutes. Each reaction was repeated one or two times for confirmation of banding positions and reproducibility. Negative controls were always used.
We initially tested 143 primers (series OPA, OPB, OPC, OPD, OPG, OPH, OPM, OPN, OPX and OPY (Operon Technologies, Alameda, CA) on An. cruzii samples to ascertain those most suited for our level of analysis. Of these, 29 generated no amplification product, 51 generated only weak bands and 63 were retested with twenty mosquito samples. The majority of the retested primer combinations produced overabundant bands that were difficult to score between samples and between gels. We therefore used only primers: OPA-01 (5'-CAGGCCCTTC-3'), OPA-11 (5'-CAATCGCCGT-3'), OPY-02 (5'-CATCGCCGCA-3'), OPY-05 (5'-GGCTGCGACA-3'), OPY-10 (5'-CAAACGTGGG-3'), OPY-15 (5'-AGTCGCCCTT-3') and OPY-16 (5'-GGGCCAATGT-3') for comparisons between samples from different populations.
The PCR products were separated on 1.5% agarose gels containing 0.5X TBE at 5V/cm during 3 hours and visualized using ethidium bromide (0.5-1.0 µg/ml). We utilized a 100-bp DNA size marker (Invitrogen). The approximate fragment size was estimated using the KODAK Digital Science EDAS 120 system.
Analysis of PCR-RAPD results. Electrophoresis profiles were compared visually and a binary table (presence-1/absence-0) created with each sample and each size position. A subsequent similarity matrix between samples was constructed using the Jaccard coefficient and a dendrogram obtained using the UPGMA (Unweighted Pair Group Method Using Arithmetic Average) method. Both the similarity matrix and dendrogram were calculated using the program NTSYS-PC (Numerical Taxonomy and Multivariate Analysis System) vers. 2.02i (Rohlf 1992).
The matrix used for calculating Analysis of Molecular Variance was prepared using AMOVA-PREP vers. 1.01 (Miller 1998). Comparisons within and between populations of An. cruzii were done using AMOVA vers. 1.5.5 (Excoffier et al. 1992). The genetic identity and genetic distance between mosquito populations and the number of migrants per generation (Nm) were calculated with POPGENE vers. 1.32 (Yeh et al. 1999) using Nei's coefficient (1972).
PCR-ITS2
Amplification of ITS2 and flanking 5.8S and 28S genes. Based on the PCR-RAPD results, 40 mosquito samples were selected for analysis of the restriction profile of ITS2 and portions of its flanking 5.8S and 28S genes. Amplification reactions were in a total volume of 25 µl containing: 1X Buffer, 3 mM MgCl2, 200 µM of each dNTP, 15 pmol of each primer (Invitrogen), 4-6 ng/µl of DNA, 1U of Taq DNA polymerase (Invitrogen) and the addition of Milli-Q sterile water to complete the final volume. Amplification cycling conditions were as follow: initial denaturing at 94ºC for five minutes, 40 amplification cycles (94ºC for two minutes, annealing at 58ºC for two minutes and extension at 72ºC for two minutes) and a final extension of 72ºC for five minutes. The primers used in this reaction were CP17 (5'-GCG CCG CGG TGT GAA CTG CAG GAC ACA TG-3') and CP16 (5'-GCG GGT ACC ATG CTT AAA TTT AGG GGG TA-3') (Mallafronte et al. 1997).
Restriction reaction. The restriction enzymes used on the amplicons of ITS2 and its flanks were determined using the An. cruzii sequences available on GenBank (accession numbers AF027165 and AF035227). Nucleotide sequences DQ386997, DQ386996, DQ364654 and DQ364653 were not available when we carried out the analyses for the study. Putative cutting sites in this species were ascertained using WebCutter 2.0 (Heiman, 1997). Eight enzymes were selected, four recognising four-base cutting sites (BstUI, HaeIII, TaqI and HhaI), two five-base sites (Sau96I and HinfI) and two six-base sites (HincII and NruI).
The restriction reaction contained 5-10 µl of the amplification product, an excess of the respective endonuclease (3-5U), 1X buffer and Milli-Q sterile water to complete a final volume of 10-20 µl. This was incubated during 3-4 hrs at the relevant temperature for each enzyme. Following the restriction reaction, fragments were separated on 2% agarose gels at 4 V/cm, and visualized using ethidium bromide and ultraviolet light. The molecular size markers used were: 1 kb Plus DNA Ladder (Invitrogen) and 123-bp DNA Ladder (Invitrogen). The restriction patterns were compared to the sequences obtained by Mallafronte et al. (1997).
RESULTS
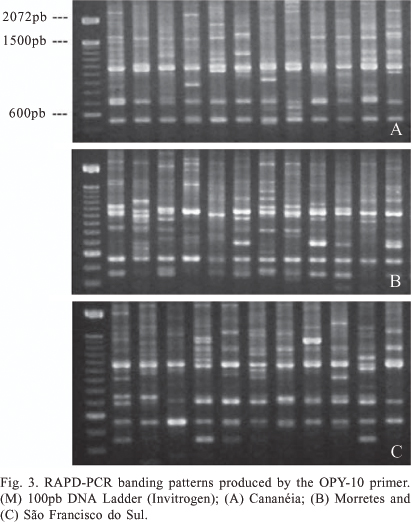
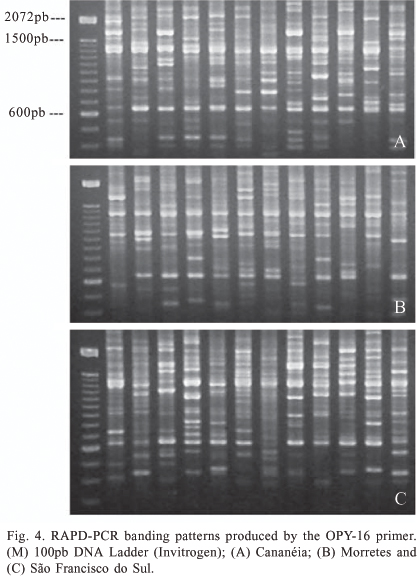
PCR-RAPD. The amplification of DNA from An. cruzii samples using PCR-RAPD revealed a high number of bands and high polymorphism, both within and between different populations. The seven primers amplified different classes of bands: monomorphic, high frequency polymorphic and polymorphic within only one or a few variant individuals. The number of these amplified fragments was different for each locality analyzed. The primers OPY-10, OPY-05 and OPY-02 yielded both monomorphic and polymorphic fragments, whilst the remainder (OPA-1, OPA-11, OPY-15 and OPY-16) amplified only polymorphic bands. The bands OPY-10, 550-bp, OPY-05, 880-bp, OPY-05, 2250-bp, OPY-05, 2550-bp, OPY-02, 1100-bp and OPY-02, 1150-bp were observed in all samples analyzed and may thus be considered species-specific molecular markers (figures 1-5).
Some fragments, whilst polymorphic, were observed in the majority of samples. Table I lists fragments with frequencies equal or greater than 90% in all samples analyzed. In addition to fragments with high frequency, some polymorphic bands were also observed that were individual-specific or observed in only two individuals. For example, of the 139 individuals analyzed, only two displayed the approximately 650-bp fragment amplified using primer OPY-10. These low-frequency polymorphisms, indicative of intraspecific variation, were found in previous work within single progeny broods (Calado & Navarro-Silva 2005).
Due to high polymorphism and variation in fragment intensities, many RAPD fragments could not be confidently compared between samples and between gels and were discarded from further analyses in the construction of the presence/absence matrix. Of all bands amplified, 59 were selected for comparison between samples using grouping analysis. Fifty-three of these were polymorphic (89.83%) and permitted the differentiation of all individuals. The similarity relationship between samples is described in figure 6. No fixed markers were observed within the same locality and UPGMA analysis also did not reveal any grouping pattern associated with geographic origin. However, some samples collected in Morretes, São Francisco do Sul and Cananéia formed a group distinct from the other samples.
Samples from Morretes amplified with primers OPA-01 and OPY-15 yielded banding patterns distinct from those of other localities. The approximately 1300-bp fragment amplified with OPA-01 was encountered in more than 80% of samples from São Paulo and Santa Catarina states but only in 17% of samples from Morretes. This was similar to the 850-bp fragment obtained using primer OPY-15, which is present in the majority of samples from São Paulo and Santa Catarina States, but absent in a large number of Morretes samples. The banding profiles from most Paranaguá and Guaratuba samples using these two primers were similar to those observed in São Francisco do Sul and Cananéia.
Analysis of Molecular Variance indicated that only 11.26% of the variation occurs between populations, with the majority (88.74%) between individuals within populations. This is apparent from direct observation of banding patterns generated by the RAPD primers, where there were several polymorphic bands between individuals of a single population. The estimated FST of 0.10 indicates a moderate amount of differentiation between populations.
Genetic distances obtained were low (0.0214-0.0673) and within the limit expected for species-level comparisons. The largest genetic distance was observed between samples from Morretes (Paraná State) and São Paulo. The values of genetic identity were high (9.350-9.788) (Table II).
The genetic similarity between populations depends primarily upon the number of migrants per generation (Nm). We estimated this parameter to evaluate the potential for dispersal from one population to another (gene flow). The mean number of migrants per generation was 4.3, indicating some gene flow between the sampled populations.
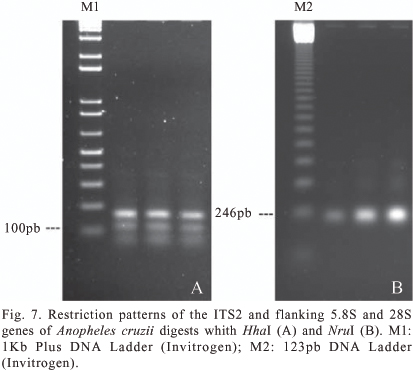
PCR-RFLP. The amplification of the ITS2 region and portions of its flanking genes (5.8S and 28S) from An. cruzii samples resulted in fragments of approximately 490-bp. Digestion with the enzymes HaeIII, HhaI, HincII, HinfII, NruI, BstUI, Sau96I and TaqI, produced 1-4 fragments that were detectable on a 2% agarose gel. The disparity observed in the sum of digest-product length and the original 5.8S-ITS2-28S amplicon was due to the presence of similar-sized fragments co-migrating or the cleavage of very small DNA fragments, which precluded fragment differentiation on 2% agarose gels. The eight endonucleases used produced fragments of similar size in all samples, with no detectable intraspecific polymorphism.
DISCUSSION
The PCR-RAPD results did not suggest the presence of more than one species in the An. cruzii complex from the samples analyzed, but do indicate that this species is highly polymorphic. The low genetic distances between samples may be related to the proximity of the collection sites, which permits genetic exchange via migration of individuals. Carvalho-Pinto & Lourenço-de-Oliveira (2004) also confirmed gene flow between populations of An. cruzii collected in the States of Rio de Janeiro, São Paulo and Santa Catarina using allozymes. These authors estimated there to be 2.99 migrants per generation and a value of Nei's genetic distance (1978) of between 0.032 and 0.083. Moreover, they noted that the populations in these States are genetically similar and represent a single species, which the results here support for populations from Cananéia (São Paulo State) and São Francisco do Sul (Santa Catarina State).
Restriction digests of the 5.8S-ITS2-28S amplicon did not reveal variation in banding patterns between our samples, although they differed from those obtained by Mallafronte et al. (1997), who confirmed the existence of two polymorphisms in An. cruzii from São Paulo State. The first resulted from the lack of an NruI restriction site in individuals collected in Peruíbe due to the deletion of two bases (AG) at positions 135/136 of ITS2. The second polymorphism, although not elaborated by the authors, comprised three nucleotides (CGC) in sequences from Peruíbe samples, which constitute part of the recognition site for the HhaI endonuclease.
The samples analyzed here showed restriction patterns compatible with sequences from São Francisco do Sul (DQ383997), Cananéia (DQ386996), Guaratuba (DQ364654) and Boracéia samples (DQ364653), but are different from those from Juquitiba (Juquitiba Municipality) (AF035227) and Peruíbe (AF027165). The samples contain the restriction site for NruI, similar to that observed by Malafronte et al. (1997) from Juquitiba samples. However, when digested with HhaI, they generated fragments of sizes more similar to those expected from the Peruíbe population (60, 162, 116, 72 and 78), than to those expected from Juquiazinho (60,277, 72 and 78) (Figure 7). These results suggest that the samples analyzed here and the six sequences available in GenBank represent at least three distinct ITS2 sequences.
Based on X chromosome banding patterns, Ramirez & Dessen (2000a,b) recognized at least three cryptic species of An. cruzii, with the following frequencies in collection localities: variant A in Boracéia (86%), Juquitiba (97%), Guaratuba (87%), Peruíbe (100%) and Cananéia (4%); variant B in São Francisco do Sul (100%), Guaratuba (13%) and Cananéia (96%); variant C in Boracéia (14%) and Juquitiba (3%).
Based on our results employing sequences avaible in GenBank, and on those presented by Ramirez & Dessen (2000a,b) and Mallafronte et al. (1997), two questions arise: are the ITS2 polymorphisms correlated to the chromosomal banding patterns? And how? One possibility is that the observed ITS2 pattern here and sequences DQ383997 (São Francisco do Sul) and DQ386996 (Cananéia) are associated to chromosomal variant B or that sequence AF027165 (Peruíbe) corresponds to variant A or that sequence AF035227 (Juquitiba) corresponds to variant C.
In São Francisco do Sul and Peruíbe, correlations can be made with greater confidence as all samples from these localities display only one chromosomal banding pattern, variants A and B, respectively. However, the number of individuals from Juquitiba that share the variant C pattern is small (3%), and thus the correlation between ITS2 and chromosomal banding pattern becomes tenuous. There is a higher probability that the sequences available from Juquitiba are related to variant A. Interestingly, in Boracéia, A and C chromosomal variants were found, however the nucleotide sequence available in GenBank that were generated from specimens collected in the same locality are similar (except at the 326 position) to those registered for São Francisco do Sul, where only the B chromosome variant was found. Similarly, for the Guaratuba (state of São Paulo) population, A chromosome variant was the most frequently observed, however the nucleotide sequence available in GenBank is similar to that found in São Francisco do Sul (B variant). Here the polymorphisms observed in ITS2 could not be correlated and thus do not represent robust taxonomic characters.
The observations made by Carvalho-Pinto & Lourenço-de-Oliveira (2004) suggest an additional possibility: that An. cruzii populations from Santa Catarina, São Paulo and Rio de Janeiro states represent a single species. These authors noted that gene flow analyses between Florianópolis (SC), Cananéia (SP), Nova Iguaçu (RJ) and Jaguaripe (BA), suggest that only the population from Bahia constituted an isolated population or a different species, whilst individuals from the other localities did not display allozyme genotypes sufficiently different to support distinct species status, as suggested by Ramirez & Dessen (2000a,b). It is worth noting that Carvalho-Pinto & Lourenço-de-Oliveira (2004) also analyzed samples from Cananéia, where two chromosomal banding variants are observed (A and B), but found no evidence for two species in this locality. A potential explanation for the homogeneity of allozyme patterns here may be the low frequency of chromosomal variant A, present in only 4% of the samples evaluated by Ramirez & Dessen (2000a,b). However the hypothesis of Carvalho-Pinto & Lourenço-de-Oliveira (2004) cannot be discarded, as these authors each used different methodologies. Moreover, although it occurs on coastal locations of the Brazilian Northeast states, An. cruzii is not considered a vector of human Plasmodium in this region (Rachou 1952). Consequently, a possible non-vector status of An. cruzii population from Northeast Brazil might support the hypothesis of two distinct species of Carvalho-Pinto & Lourenço-de-Oliveira (2004). Thus, if the populations present in the Southern and Southeastern regions comprise one species, the conserved polymorphisms in ITS2 combined with variable chromosome banding patterns may be indicative of An. cruzii genetic diversity only and have no taxonomic significance.
The data obtained until present through cytogenetic and molecular studies, although conflicting, do suggest that the An. cruzii taxon is highly polymorphic and that it potentially constitutes a species complex. However, there is still no consensus as to the geographic distribution of its component species. Neither is there agreement as to the proper methodology to correctly ascertain this information. Studies involving characters (morphological, genetic or biochemical) between species of Anopheles are sometimes ambiguous because of the limited distributions of some species. Consequently, these characters may become uninformative when applied to the entire geographic distribution of a taxon (Fritz et al. 1991; Fritz et al. 1995). In addition, chromosomal inversions and molecular markers are not always congruent for interspecific discrimination within the Anopheles genus (Dusfour et al. 2004). Thus, the possibility that An. cruzii comprises a species complex deserves additional investigations using a larger number of samples, collections sites and additional methodologies that permit efficient comparisons with research already published. Additional ITS2 loci must be sequenced to determine if polymorphisms are shared between localities or if they are relegated to specific regions and to verify if the chromosomal banding patterns defined by Ramirez & Dessen (2000a,b) are correlated to the fixed polymorphisms of ITS2 and/or other regions or genes.
Acknowledgements. We are indebted to Dr. Luiz Gonzaga Esteves Vieira of the Plant Biotechnology Laboratory of the Paraná State Agronomy Institute for the use of the molecular laboratory. We wish to also acknowledge the Paraná State Environmental Institute for the collection license (number 01/02) in the Ilha do Mel Ecological Station; to CNPq for support with PhD funding and bench fees for DCC and research productivity funding to MANS. We also thanks Pedro MC Pedro for his help in the translation. MAMS and MANS are financially supported by FAPESP grant number 05/53973-0.
Received 13/06/2006; accepted 28/08/2006
- Branquinho, M. S.; M. T. Marrelli; I. Curado; D. Natal; J. M. S. Barata; R. Tubaki; G. C. Carréri-Bruno; R. T. Menezes & J. K. Kloetzel. 1997. Infecção do Anopheles (Kerteszia) cruzii por Plasmodium vivax e Plasmodium vivax variante VK247 nos municípios de São Vicente e Juquitiba, São Paulo. Revista Panamericana de Salud Publica 2: 189193.
- Calado, D. C. & M. A. Navarro-Silva. 2005. Polimorfismos em Anopheles (Kerteszia) cruzii Dyar & Knab (Diptera, Culicidae) detectados através de marcadores PCR-RAPD: Variabilidade genética entre indivíduos de mesma progênie. Revista Brasileira de Zoologia 22: 11211126.
- Carvalho, A. O. & L. G. E. Vieira. 2001. Determinação das condições ótimas para análise de PCR-RAPD em Atta sexdens rubropilosa Forel (Hymenoptera: Formicidae). Neotropical Entomology 30: 593600.
- Carvalho-Pinto, C. J. & R. Lourenço-de-Oliveira. 2003. Izoenzimatic analysis of four Anopheles (Kerteszia) bellator Dyar & Knab (Diptera: Culicidae) populations. Memórias do Instituto Oswaldo Cruz 98: 10451048.
- Carvalho-Pinto, C. J. & R. Lourenço-de-Oliveira. 2004. Isoenzimatic analysis of four Anopheles (Kerteszia) cruzii (Diptera: Culicidae) populations of Brazil. Memórias do Instituto Oswaldo Cruz 99: 471475.
- Cheung, W. Y.; N. Hubert & B. S. Landry. 1993. A simple and rapid DNA microextraction method for plant, animal and insect suitable for RAPD and other PCR analyses. PCR Methods and Applications 3: 6970.
- Coutinho, J. O.; R. Rachou & I. Ricciardi. 1943. Contribuição para o conhecimento dos transmissores de malária no Brasil A. (K.) cruzi e A. (K) bellator. Arquivos de Higiene 13: 115127.
- Dusfour, I.; Y-M. Linton; A. Couhet; R. E. Harbach; V. Baimai; H. D. Trung; C. M. Seng; A. Matusop & S. Manguin. 2004. Molecular evidence of speciation between island and continental populations of Anopheles (Cellia) sundaicus (Diptera: Culicidae), a principal malaria vector taxon in Southeast Asia. Journal of Medical Entomology 41: 287295.
- Excoffier, L.; P. E. Smouse & J. M. Qattro. 1992. Analysis of molecular variance inferred from metric distance among DNA haplotypes application to human mitochondrial DNA restriction sites. Genetics 131: 479491.
- Fritz, G. N.; S. K. Narang; D. L. Kline; J. A. Seawhight; R. K. Washino; C. H. Porter & F. H. Collins. 1991. Diagnostic characterization of Anopheles freeborni and An. hermsi by hybrid crosses, frequencies of polytene X chromosomes and rDNA restriction enzyme fragments. Journal American Mosquito Control Association 7: 198206.
- Fritz, G. N.; H. Bermudez & J. A. Seawright. 1995. Genetic differentiation and diagnostic loci of Anopheles nuneztovari, An. trikae and An. rangeli (Diptera: Culicidae). Journal of Medical Entomology 32: 663672.
- Heyman, M. 1997. Webcutter version 2.0 Available at: http://www.firstmarket.com /cutter/cut2.html Last access: March 2005.
- Mallafronte, R. S.; M. T. Marrelli; G. C. Carreri-Bruno; P. R. Urbinatti & O. Marinotti. 1997. Polymorphism in the second internal spacer (ITS2) of Anopheles (Kerteszia) cruzii (Diptera: Culicidae) from the State of São Paulo, Brazil. Memórias do Instituto Oswaldo Cruz 92: 306.
- Miller, M. P. 1998. AMOVA-PREP versão 1.01. A program for the preparation of AMOVA input files from dominat-marker row data. Departamento of Biological Sciences, Northern Arizona University, AZ. Disponível em: http://www.marksgeneticsoftware.net Último acesso em: março de 2005.
- Nei, M. 1972. Genetic distance between populations. American Naturalist 106: 283292.
- Nei, M. 1978. Estimation of average heterozigosity and genetic distance from a small number of individuals. Genetics 89: 583590.
- Rachou, R. G. 1952. Sobre o combate aos anofelinos do subgênero Kerteszia no sul do Brasil. Revista Brasileira de Malariologia e Doenças Tropicais 4: 245254.
- Ramirez, C. C. L. & E. M. B. Dessen. 2000a. Chromosomal evidence for sibling species of the malaria vector Anopheles cruzii Genome 43: 143151.
- Ramirez, C. C. L. & E. M. B. Dessen. 2000b. Chromosome differentiated populations of Anopheles cruzii: evidence for a third sibling species. Genetica 108: 7380.
- Rohlf, F. J. 1992. NTSYS-PC. Numerical taxonomy and multivariate analysis systems. Exeter Software, Applied Biostatistics, N. Y. 225pp.
- Yeh, F. C.; T. J. B. Boyle; Y. Z. Ye & J. M. Xiyan. 1999. POPGENE version 1.32. Microsoft Window-based freeware for population genetics analysis. University of Alberta and Center for International Forestry Research. Available at http://www.ualberta.ca/~fyeh/index.htm Last access: March 2005.
Publication Dates
-
Publication in this collection
27 Oct 2006 -
Date of issue
Sept 2006
History
-
Received
13 June 2006 -
Accepted
28 Aug 2006