Abstracts
Translatable and nontranslatable versions of the coat protein (cp) gene of a Papaya ringspot virus (PRSV) isolate collected in the state of Bahia, Brazil, were engineered for expression in Sunrise and Sunset Solo varieties of papaya (Carica papaya). The biolistic system was used to transform secondary somatic embryo cultures derived from immature zygotic embryos. Fifty-four transgenic lines, 26 translatable and 28 nontranslatable gene versions, were regenerated, with a transformation efficiency of 2.7%. Inoculation of cloned R0 plants with PRSV BR, PRSV HA or PRSV TH, Brazilian, Hawaiian and Thai isolates, respectively, revealed lines with mono-, double-, and triple-resistance. After molecular analysis and a preliminary agronomic evaluation, 13 R1 and R2 populations were incorporated into the papaya-breeding program at Embrapa Cassava and Tropical Fruits, in Cruz das Almas, Bahia, Brazil.
Carica papaya; potyvirus; resistance; breeding; virus
Versões traduzíveis e não traduzíveis do gene da capa protéica (cp) de um isolado de Papaya ringspot virus (PRSV) coletado no Estado da Bahia, Brasil, foram produzidas para expressão nas variedades Sunrise e Sunset Solo de mamoeiro (Carica papaya). O sistema de biobalística foi utilizado para transformar embriões somáticos secundários derivados de embriões zigóticos imaturos. Cinqüenta e quatro linhas transgênicas, sendo 26 contendo versões traduzíveis e 28 contendo versões não traduzíveis do gene cp foram regeneradas, o que resultou em 2,7% de eficiência de transformação, quando considerado o número de linhas transgênicas obtidas por embrião zigótico imaturo excisado. Desafios de plantas R0 com PRSV BR, PRSV HA ou PRSV TH, respectivamente isolado brasileiro, havaiano e tailandês, revelaram linhas com resistência a um, dois e três isolados de PRSV. Após análises moleculares e avaliação agronômica preliminar, 13 populações R1 e R2 de mamoeiros transgênicos foram incorporadas ao programa de melhoramento genético da Embrapa Mandioca e Fruticultura, em Cruz das Almas, Bahia, Brasil.
Carica papaya; potyvirus; resistência; melhoramento; PRSV
ARTICLES ARTIGOS
Development of virus resistant transgenic papayas expressing the coat protein gene from a Brazilian isolate of Papaya ringspot virus
Desenvolvimento de mamoeiros transgênicos resistentes a vírus expressando o gene da capa protéica de um isolado brasileiro de Papaya ringspot virus
Manoel T. Souza JúniorI; Osmar NickelII; Dennis GonsalvesIII
IEmbrapa Genetic Resources and Biotechnology, Caixa Postal 02372, Brasília, DF, CEP 70770-900, Brazil, e-mail:msouza@cenargen.embrapa.br
IIEmbrapa Grapes & Wine, Cx.Postal 130, Bento Gonsalves, RS, CEP 95700-000, Brazil
IIIDepartment of Plant Pathology, Barton Lab., NYSAES, Cornell University, 14.456, Geneva, NY, USA
ABSTRACT
Translatable and nontranslatable versions of the coat protein (cp) gene of a Papaya ringspot virus (PRSV) isolate collected in the state of Bahia, Brazil, were engineered for expression in Sunrise and Sunset Solo varieties of papaya (Carica papaya). The biolistic system was used to transform secondary somatic embryo cultures derived from immature zygotic embryos. Fifty-four transgenic lines, 26 translatable and 28 nontranslatable gene versions, were regenerated, with a transformation efficiency of 2.7%. Inoculation of cloned R0 plants with PRSV BR, PRSV HA or PRSV TH, Brazilian, Hawaiian and Thai isolates, respectively, revealed lines with mono-, double-, and triple-resistance. After molecular analysis and a preliminary agronomic evaluation, 13 R1 and R2 populations were incorporated into the papaya-breeding program at Embrapa Cassava and Tropical Fruits, in Cruz das Almas, Bahia, Brazil.
Additional keywords: Carica papaya, potyvirus, resistance, breeding, virus.
RESUMO
Versões traduzíveis e não traduzíveis do gene da capa protéica (cp) de um isolado de Papaya ringspot virus (PRSV) coletado no Estado da Bahia, Brasil, foram produzidas para expressão nas variedades Sunrise e Sunset Solo de mamoeiro (Carica papaya). O sistema de biobalística foi utilizado para transformar embriões somáticos secundários derivados de embriões zigóticos imaturos. Cinqüenta e quatro linhas transgênicas, sendo 26 contendo versões traduzíveis e 28 contendo versões não traduzíveis do gene cp foram regeneradas, o que resultou em 2,7% de eficiência de transformação, quando considerado o número de linhas transgênicas obtidas por embrião zigótico imaturo excisado. Desafios de plantas R0 com PRSV BR, PRSV HA ou PRSV TH, respectivamente isolado brasileiro, havaiano e tailandês, revelaram linhas com resistência a um, dois e três isolados de PRSV. Após análises moleculares e avaliação agronômica preliminar, 13 populações R1 e R2 de mamoeiros transgênicos foram incorporadas ao programa de melhoramento genético da Embrapa Mandioca e Fruticultura, em Cruz das Almas, Bahia, Brasil.
Palavras-chave adicionais: Carica papaya, potyvirus, resistência, melhoramento, PRSV.
INTRODUCTION
Papaya (Carica papaya L.) is the most cultivated species of the Caricaceae family. It is grown in lowland tropical and subtropical areas around the world, and is usually consumed as fresh dessert fruit (Manshardt, 1992).
Papaya ringspot is the most important disease limiting the production of papaya worldwide. It is characterized by severe leaf mosaic, reduction of leaf canopy as the disease progresses, stunting of the plant, water-soaked oily streaks on the petiole and upper part of the trunk, and ringed spots on the surface of the fruits. Papaya ringspot virus (PRSV), family Potyviridae, genus Potyvirus, strain P, is the causal agent of this disease, and it is transmitted by several species of aphids to papaya and members of the Cucurbitaceae family in a non-persistent manner. The PRSV has flexuous rod-shaped particles of 780x12 nm and a genome consisting of a linear positive single stranded RNA (Purcifull et al., 1996) of approximately 10 kb (Yeh et al., 1992). The PRSV was first reported in Brazil in 1969 (Costa et al., 1969). Nowadays, PRSV is found in almost all papaya production areas in Brazil, causing losses of up to 70% (Barbosa & Paguio, 1982).
The development of plant transformation systems and the formulation of the parasite-derived resistance (PDR) concept (Sanford & Johnson, 1985), offered an additional tool to control virus diseases. In 1992, Fitch et al. (1992) reported on virus resistant papaya, containing the translatable version of the coat protein (cp) gene of PRSV HA 5-1, leading to PRSV-resistant papayas (Manshardt, 1998; Gonsalves, 1998).
In this study we report on the development of transgenic papayas with resistance to Brazilian PRSV isolates. Translatable and untranslatable PRSV BR cp genes were cloned and used to transform somatic embryos of the Sunrise and Sunset Solo varieties by means of the Biolistic-mediated transformation system.
MATERIAL AND METHODS
Papaya ringspot virus isolates, primers, and cp gene RT-PCR amplification and cloning
Three geographically different PRSV isolates were used in this study: 1) a Brazilian isolate (PRSV BR) collected in the southeast region of the State of Bahia; 2) a Hawaiian isolate (PRSV HA) collected from the island of Oahu, Hawaii in 1977 (Gonsalves & Ishii, 1980); and 3) an isolate from Thailand (PRSV TH) collected in Khon Kaen (Tennant et al., 1994). The PRSV BR was used as template for the amplification of the cp gene versions by RT-PCR, and for the challenge of R0 and R1 plants. All isolates were maintained in the greenhouse in papaya and Cucumis metuliferus L.
Three different versions of the PRSV BR cp gene were amplified and cloned. The primers 5'-ATCATTCCATGGCT GTGGATGCTGGTTTG-3' and 5'-AGCTAACCATGGGGT GAAACAGGGTCG-3' were used as 5'-end and 3'-end primers, respectively, to amplify the Translatable Short (TS) version, the primers 5'-ATCATTCCATGGTCCAAGAATGA AGCTGT-3' and 5'-AGCTAACCATGGGGTGAAACAGG GTCG-3' to amplify the Untranslatable Medium (UM) version, and the primers 5'-ATCATTCCATGGGCGTGTTC CATGAATCAA-3' and 5'-AGCTAACCATGGGCGAGTAT TCAGTTGCGC-3' to amplify the Translatable Long (TL) version.
Total plant RNA from PRSV BR infected papaya plants was extracted as described by Napoli et al. (1990). The reverse transcription was done under the following conditions: 1-2 µg of total RNA, 200 ng of 3'-end primer, 200 ng of each dNTP, 10 mM DTT, 80 units of RNAsin, 360 mM of 2-mercaptoethanol, 1X RT buffer, and 400 units of M-MLV RT (Maloney Murine Leukemia virus) (Promega, Madison, WI). Initially, a 15-µl aliquot containing only the total RNA and the 3'-end primer was heated at 70 ºC for 5 min, and cooled on ice for 2 min. Then, a 35-µl aliquot containing the other reaction components was added and incubated for 90 min at 37 ºC. Incubating at 70 ºC for 5 min stopped the reaction.
After the RT reaction, 5 µl of RT solution with single strand cDNAs was used as template for PCR under the following conditions: 100 ng of each dNTP, 1X PCR buffer, 100 ng of each 5'-end and 3'-end primers, 1.5 mM MgCl2, and 2.5 units of Taq DNA polymerase (Perkin-Elmer Corp., Norwalk, CT) per tube, in a 50 µl volume. A cycle of 94 ºC/ 3 min, 40 ºC/ 1 min, and 72 ºC/ 3 min, was followed by 30 cycles of 92 ºC/ 1 min, 53 ºC/ 1 min, and 72ºC/ 2.5 min, and by one cycle of 72 ºC/ 7 min. The same conditions were used to amplify all cp gene versions. All PCR products were analyzed in 1% agarose gel electrophoresis buffered in 1X TAE (Sambrook et al., 1991) and stained with ethidium bromide, except as otherwise stated.
All oligonucleotide primers used for the amplification of the cp gene versions were designed containing the Nco I restriction site, chosen to insert the RT-PCR products in the pEPT8 (Jan, 1998) vector. The Hind III restriction site was chosen to excise the cp gene-containing EPT8 cassette from the vector pEPT8, and to insert it into the binary vector pGA482G (Jan, 1998), derived from the cosmid vector pGA482 (An, 1986), with npt gene as the plant selectable marker. The plant expression cassette EPT8 is 884 pair-long bases, and has a double 35S enhancer, the 35S promoter, the Alfalfa mosaic virus (AlMV) leader sequence, a multiple cloning site (MCS), and the 35S terminator (Jan, 1998).
Papaya material, tissue culture and transformation, selection and maturation of transgenic embryos, and acclimatizing of plantlets
Somatic embryogenic cultures were derived from immature zygotic embryos from the Hawaiian Solo papaya cultivars Sunrise and Sunset. The zygotic embryos were excised from seeds harvested approximately 100 days after anthesis (Fitch et al., 1990). The fruits were surface sterilized for one hour in 20% Clorox/ 0.1% Tween 20, and the seeds removed.
A papaya tissue culture system (Fitch et al., 1990; Fitch, 1993), modified by Cai et al. (1999), was generally followed. Plasmid DNA (3 µg/plate) was transferred into embryos using a helium-driven biolistic apparatus. Procedures for microprojectile coating and loading were conducted as described by Sanford et al. (1993).
Bombarded papaya cultures were transferred to fresh induction medium on the same day of the bombardment or one day later. These cultures were kept at 28 ºC in the dark for seven-ten days and then transferred to induction medium supplemented with kanamycin as selection agent. Putative transgenic embryo clusters (PTECs) were selected on induction medium (IM) supplemented with 150 mg/l of kanamycin, in ten rounds of sub culturing and selection, and then transferred to maturation medium MM (IM without 2,4-D and kanamycin, supplemented with 250 mg/l carbenicillin and 0.1% activated charcoal). The PTECs were incubated under cool white fluorescent lights 70-90 µE/ m2/ second for a 12 h photoperiod at 25 ºC for two-four weeks. Embryos were considered mature after further development of cotyledonary structures and greening of the material.
Mature embryos were transferred to germination medium (GM) supplemented with 250 mg/l of carbenicillin, and maintained under the same conditions as for maturation. Transfer to fresh GM was done on a monthly basis. Embryos were considered germinated as soon as true leaves and radicles were observed.
Small plantlets (1-2 cm height) were transferred to jars with 30 ml of sterile vermiculite and 25 ml of liquid GM, incubated for maturation and germination for up to one month. Vigorously growing plants (two-five leaves, 2-5 cm stem, and a 2-15 cm tap root) were removed from jars, potted (10x10 cm) in a soil:vermiculite:perlite mixture (1:1:1 by volume), covered with a clear plastic bag, and transferred to the greenhouse. After one week small holes were punctured in the bags and they were gradually enlarged every few days to acclimatize plants to normal greenhouse conditions. The bags were completely removed after two-three weeks.
Analysis of transgenic plants
PCR, Southern blot and Elisa analyses: Plant genomic DNA of transgenic and nontransgenic papaya plants was extracted as follows: 1) 750 µl of extraction buffer [one volume of DNA extraction buffer (0.35 M Sorbitol, 0.1 M Tris, 5 mM EDTA, 25 mM Sodium bisulfite, pH 8.26), 1 volume nuclei lysis buffer (0.2 M Tris, 50 mM EDTA, 2 M NaCl, 2% CTAB), and 0.4 volume of 5% Sarkosyl] was added to a 2 ml micro tube containing 100-200 mg of leaf tissue ground in liquid N, and then incubated at 65 ºC for 30-60 min; 2) 750 µl of chloroform:isoamyl alcohol (24:1) was added to the mix and centrifuged at 10,000 rpm for 10 min; 3) the upper layer was removed and mixed with one volume of cold isopropanol, and then centrifuged at 10,000 rpm for 10 min; 4) the pellet was rinsed with cold 75% ethanol, dried for 5 min in speed vacuum, ressuspended in 50 µl of TE buffer (pH 7.5) (Sambrook et al., 1991), and then treated with 10 µl of RNase A (1 mg/ml) at 37 ºC for 30 min. Quantified DNA was stored at -20 ºC until assaying the presence of the cp transgene by PCR.
A 2 µl aliquot with 100 ng/ µl solution of genomic DNA was used as template for PCR under the following conditions: 100 ng of each dNTP, 1X PCR buffer, 100 ng of each 5'-end and 3'-end primers, 1.5 mM MgCl2, and 2.5 units of Taq DNA polymerase per tube, in a 50 µl volume. A cycle of 94 ºC/ 3 min, 40 ºC/ 1 min, and 72 ºC/ 3 min, was followed by 30 cycles of 92 ºC/ 1 min, 53 ºC/ 1 min, and 72 ºC/ 2.5 min, and by a cycle of 72 ºC/ 7 min. The same conditions were used to amplify all cp gene constructs, and the PCR products were separated as described above.
Ten micrograms of Hind III-digested DNA were separated by electrophoresis in 1% agarose gel buffered in 1X TBE, stained with ethidium bromide, and transferred to a GeneScreenPlus nylon membrane (BioTechnology Systems, NEN Research Products, Boston, MA) using capillary transfer as described in the manufacturer's protocol. The membrane was hybridized with a probe consisting of the cp gene excised from the TL version of the gene described earlier in this work, and labeled with 32P by using a random primer based system (Feinberg & Vogelstein, 1983).
Papaya leaves were homogenized in extraction buffer (0.25 M potassium phosphate, 0.1 M EDTA, pH 7.5) (Gonsalves & Ishii, 1980) and then centrifuged at 5,000 rpm at 4ºC for 3 min. Total protein was quantified in the sample by the Bio-Rad Protein Assay (Bio-Rad Laboratories, NY).
The npt II gene expression (Cabanes-Bastos et al., 1989) in plant was detected by Enzyme linked immuno absorbant assay (Elisa) according to the manufacturer's conditions (5 Prime-3 Prime Inc., Boulder, CO). The samples tested were 100 µl of crude sap processed as above and adjusted to contain 100 µg of total protein. Dilutions of 1:1000, 1:2000, and 1:5000 were used for anti-npt II antibody, biotinylated anti-NPT II antibody, and streptavidin conjugated alkaline phosphatase, respectively. The absorbance was measured at 405 nm with a MicroELISA AutoReader MR700 (Dynatech Inc., Chantilly, VA) 15 min after the addition of paranitrophenyl phosphate (1 mg/ml, 10% diethanolamine, pH 9.8).
Double antibody sandwich Elisa (DAS-Ellisa) was used to measure cp levels in tissue extracts containing 100 µg of total protein as measured by the Bio-Rad Protein Assay. Polyclonal and monoclonal antibodies, produced against PRSV HA 5-1, were used for coating and as a conjugate, respectively. The reaction was measured at 405 nm as before at different times after the addition of p-nitrophenyl phosphate (1 mg/ml, 10% diethanolamine, pH 9.8).
Challenge of transgenic papayas by PRSV isolates in the greenhouse: PRSV HA, PRSV BR, and PRSV TH were used to challenge R0 and R1 transgenic plants containing the cp gene of the Brazilian PRSV isolate. After being dusted with carborundum powder, transgenic and nontransgenic plants having ten-15 (R0 plants) or six-ten (R1 plants) true leaves were rubbed with inoculum (1:20, w/v) diluted in cold 0.01 M potassium phosphate buffer (pH 7.5). Two leaves were inoculated per plant, and subsequently rinsed with water.
All inoculated plants were observed weekly for three weeks. Plants that remained symptomless were re-inoculated and kept under observation for three more weeks. Disease resistance was assessed on new growth of transgenic and nontransgenic plants by comparing the rate of symptom development (vein clearing, mosaic, leaf distortion, leaf reduction) and the severity of symptoms.
R1 seeds were treated in 1 M potassium nitrate solution for 30 min under constant agitation (Nagao & Furutani, 1986), and then further treated with Captan (0.1% in 1M KNO3) for 2-5 min. Seeds were allowed to germinate in a soil:vermiculite:perlite mixture (1:1:1 by volume). The seedlings were transplanted to pots containing the same soil mixture, and kept in the greenhouse for the duration of the experiment.
Field trial with transgenic papayas: A field trial was begun at Embrapa Genetic Resources and Biotechnology, Brasília, DF, Brazil, in January 2000 (CTNBio process nº 01200.003364/99-99), and terminated in July 2001. Transgenic papayas were cultivated mainly for seed production and preliminary evaluation of agronomic traits, such as plant sex, fruit shape and color of the flesh. Controlled crosses were performed on female and hermaphrodite papayas. All hermaphrodite flowers not used for crossing were discarded before the opening of the flower, in order to reduce pollen release in the environment. All fruits originated from controlled crosses were harvested before maturation to avoid attack by insects, birds and bats. Dry seeds were stored at 4 ºC or 20 ºC until further use.
RESULTS
Translatable and untranslatable versions of the coat protein gene
All three versions of the cp gene of PRSV BR were obtained. Including the stop codon, the RT-PCR product of the translatable short (TS) version had the 849 nucleotides at the 3'-end of the cp gene, and the untranslatable medium (UM) had 862. The translatable long construct had all 921 nucleotides of the PRSV BR cp gene (Souza Jr., 1999). The long cp gene has its 5'-end starting at Yeh's start site (Yeh et al., 1992), while both medium and short cp gene have it from the Quemada's start site (Quemada et al., 1990).
Embryo excision and somatic embryogenesis
Two thousand immature zygotic embryos (Figure 1A) were excised from three batches of papaya fruits (one of 'Sunset' and two of 'Sunrise'). Primary somatic embryos started to develop on the apical dome of the zygotic embryo as early as one week after the excision (Figure 1B). After four weeks, over 50% of the zygotic embryos had generated somatic embryos that were suitable for sub-culturing (Figure 1C and 1D). The primary and secondary embryos developed through direct somatic embryogenesis; no callus stage was observed. Somatic embryos were sub-cultured on fresh induction medium for further secondary embryo initiation and multiplication. The subculture phase took two to four months, and was done in order to produce sufficient masses of embryos to be bombarded with the several cp constructs.
Transformation and selection of putative transgenic embryo clusters
Six transformation experiments were done using all cp construct versions. Experiments 1, 3a, 3b, and 3c used Sunrise somatic embryos, while experiments 2a and 2b used Sunset (Table 1). In experiment 1, eight plates were bombarded with the TL version or the cp gene, sixteen with UM, and eight with tungsten particles only. In experiment 2a, seven plates were bombarded with TL, seven with UM, and four with tungsten particles only. In experiment 2b, six plates were bombarded with TL and six with UM; the same was done in experiment 3a. In experiment 3b, nine plates were bombarded with TS. In the last experiment, 3c, five plates were bombarded with TL, five with UM, and five with TS. In each one of the six experiments, two to four plates were kept as no-bombardment controls. A total of 86 plates were bombarded; however, eight plates were lost because of problems with contamination by fungi and/or bacteria. Data in Table 1 do not consider the plates that were discarded.
Putative transgenic embryo clusters, hereafter designated as PTECs (Figure 1E) appeared as early as two months after bombardment. However, in some experiments it took up to six months after the bombardment to select the first PTEC (Table 1). Each one of the six experiments was submitted to ten rounds of selection on a monthly basis, starting in the second month after the bombardment. A total of 429 PTECs were selected, giving an average number of 5.5 per plate. A total of 17 out of 78 plates (22%) that underwent selection did not produce PTEC. The highest number of PTEC selected per plate was 24. None of the non-bombarded controls or those that were bombarbed with tungsten particles only developed viable embryo cluster under selection conditions.
Maturation and plant regeneration of putative transgenic embryo clusters
In general, three to four weeks on maturation medium were enough for most embryos to develop to the torpedo or cotyledonary stage (Figure 1F, 1G). Although some embryo clusters were not at the globular stage after one month on maturation medium, they were also transferred to germination medium. After transfer, the PTECs were sub-cultured on a monthly basis. In a few cases, spontaneous development of callus from PTECs cultured on germination medium was observed (Figure 1H). For most of the PTECs, as the germination stage progressed, some embryo clusters turned brown and were removed from the petri dish to provide more space for those that were germinating (Figure 2A and 2B).
All PTECs that generated cotyledonary embryos also developed roots on germination medium (Figure 2C). Regenerated plantlets were transferred to baby food jars containing liquid germination medium and vermiculite (Figure 2D). Plantlets with shoots and roots about 1 cm long grew well in the germination medium and vermiculite, and were ready for transfer to soil after three-four weeks (Figure 2E and 2F). Nearly all the plants that were transferred to soil and acclimated in the greenhouse survived and grew rapidly once the plastic bags were removed.
A total of 54 putative transgenic lines were regenerated from 429 PTECs, giving a plant regeneration efficiency rate of 12.6%. Thus, as 2,000 zygotic embryos were excised in the beginning of the work, an average of one transgenic line was obtained out of each group of 37 zygotic embryos excised, giving a 2.7% putative transformation rate. At the end of the regeneration phase, 67% of the PTECs, which had regenerated to plants, produced two or more plants. Plants from the same PTEC were considered as clones of one line. Previous Southern blot studies (Cai et al., 1999) showed that this was true in 100% of the cases. Putative transgenic lines (PTLs) were obtained from 34 different bombarded plates, with some plates producing up to six lines. Two PTLs died in the greenhouse before molecular characterization.
Molecular characterization and PRSV challenge of R0 putative transgenic plants
Each PTL was tested by PCR in order to detect the cp gene, by DAS-Elisa to detect the coat protein, and by Elisa analysis to detect the NPT II enzyme. Eight (15.4%) of the 52 lines tested were PCR negative for the cp gene (data not shown). The CP levels of the PTLs were compared to Rainbow or SunUp, which are transgenic plants known to express the cp gene (Gonsalves, 1998), and to healthy nontransgenic papayas, by DAS-Elisa analysis. The CP Elisa readings of most of the lines were similar to nontransgenic papayas, and only five of the 52 lines had readings two or more times higher than Rainbow or SunUp (data not shown). When tested by Elisa, the NPT II readings of all PTLs were two or more times higher than nontransgenic papayas, revealing that all of them were transformants (data not shown). These results suggest that the plants PCR negative for the cp gene, but positive for NPT II expression, somehow probably had the cp gene disrupted during its integration in the papaya genome, making it unfeasible for identification by PCR.
The PTLs with only one plant were not challenged as R0 plants; instead, they were grown to produce R1 seeds. For lines with only two plants, a plant was saved as back up, and the other challenged by the homologous isolate PRSV BR. Those with three plants had a plant saved, one challenged by the homologous isolate, and the other challenged by the heterologous isolate PRSV HA. Those with four or more plants had a plant saved, one challenged by the homologous isolate, another challenged by PRSV HA, and the fourth challenged by PRSV TH. Plants that had not developed any symptoms three weeks after the first mechanical inoculation were re-inoculated. Plants not showing symptoms three weeks after the second inoculation were classified as resistant. All resistant plants were kept longer in order to see if breakdown of the resistance could happen later.
Inoculated plant showed a range of phenotypes. Some plants showed symptoms similar to the symptoms seen in the non-transformed control plants infected by the virus, while others showed a one to two week delay in symptom appearance. However, the delayed symptoms were as severe as those observed in infected non-transformed plants. These plants were considered susceptible to PRSV. Another group of plants showed a much longer delay in symptom appearance (more than six weeks from the first inoculation). The plants initially had milder symptoms, which became more severe as the plants grew. These plants were considered as tolerant to PRSV. Finally, some plants did not show any symptoms, revealing an apparent immunity to the virus. These plants showing no symptoms were considered as resistant to PRSV. As expected, those lines negative for the presence of the cp gene were susceptible to PRSV infection.
Twenty-nine of the 52 R0 lines obtained were challenged with the Brazilian isolate of PRSV; 11 (38%) were resistant, three tolerant, and 15 susceptible. Twenty-six lines of the 29 challenged by the homologous isolate were also challenged with PRSV HA; resulting in seven resistant (27%), two tolerant, and 17 susceptible. Twenty-two of the 26 challenged by the homologous isolate and by the Hawaiian isolate were also challenged with PRSV TH, resulting in two resistant (9%), one tolerant, and 19 susceptible.
Molecular and agronomic characterization, and PRSV challenge of R1 and R2 populations
The R1 progenies of three R0 lines, obtained by self-crosses, were screened for the presence of the cp gene and for resistance to PRSV from Brazil, Hawaii, and Thailand in the greenhouse (Table 2). UM6g, TL2b and UM1a R0 plants were positive for the presence of the cp gene when assayed by PCR, while their progenies segregated for the presence of the cp gene (UM stands for nontranslatable medium - this plant was transformed with the UM version of the cp gene, while 6g indicates that this line was regenerated from the seventh PTEC selected in the sixth plate transformed with that cp gene version; TL stands for translatable large). The PCR assay revealed that 47 UM6g R1 plants out of 60 tested positive for the presence of the cp gene, suggesting a 3:1 segregation ratio for this gene. The same was true for theUM1a R1 population, where 19 out of 23 R1 plants were also positive for the presence cp gene. In the case of the TL2b R1 population, 23 out of the 45 plants were cp positive, which gave a 1:1 segregation ratio.
The UM6g R0 plants were susceptible to all three isolates, while TL2b R0 plants were resistant only to the Brazilian isolate, and UM1a R0 plants were resistant to all three isolates. In the case of their R1 progenies, all cp UM6g and TL2b R1 plants, positive by PCR, were susceptible to all three PRSV isolates (Table 2). Interestingly, the TL2b R1 plants cp positive by PCR did not maintain the resistance to the homologous isolate seen in the TL2b R0 plant. In the case of UM1a R1 plants, all were resistant to the homologous isolate as well as to the Hawaiian isolate, and only two of the seven plants challenged by the isolate from Thailand were resistant to the virus.
Another R1 population, from a self-cross of the TS1j R0 plant (TS stands for translatable short), and a R2 population, from a selfed R1 PRSV resistant plant, which had been obtained from the cross between UM7c e UM1g R0 plants, were also screened for the presence of the cp gene by PCR and for resistance to other PRSV isolates from Brazil (PRSV BA-CA from Cruz das Almas, Bahia, and PRSV DF from Brasília, Distrito Federal) (Lima et al., 2002). TS1j and UM1g R0 plants have not been challenged by PRSV, and UM7c R0 plant were resistant to PRSV BR. The PRSV cp gene segregated at an approximately 3:1 ratio (Figure 3), and resistant plants were observed in both progenies.
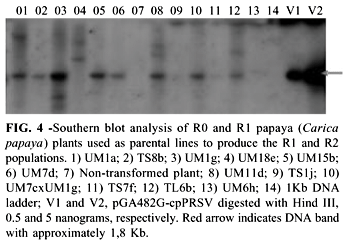
Based on the molecular characterization and the preliminary agronomic evaluation done during the field trial, a group of 13 R1 and R2 progenies were selected and their seeds, which were produced under controlled pollination, were incorporated into the papaya breeding program at Embrapa Cassava and Tropical Fruits, in Cruz das Almas, Bahia (Table 3). The R0 plants that generated these populations were all positive for the presence of the cp gene, showing one or more insertions of this gene (Figure 4). Twelve of these progenies were challenged in the greenhouse for resistance to PRSV BA-CA and all of them showed plants resistant to this isolate (Filho et al., 2004).
DISCUSSION
The major objective of this study was to obtain transgenic Sunrise and/or Sunset plants that were resistant to the tested PRSV isolates from Brazil. These plants will be used in breeding programs as source of resistance to generate papaya varieties or hybrids with a broad spectrum of resistance to this virus. Our results showed that some transgenic papaya plants expressing the cp gene of a Brazilian isolate of PRSV are resistant to mechanical inoculation with this virus isolate, as well as other Brazilian isolates. Some of these plants were also resistant to a Hawaiian and an isolate from Thailand.
A total of 54 different transgenic lines (26 translatable and 28 untranslatable) were produced, giving a transformation efficiency of 2.7% and a regeneration efficiency of 12.6%. As pointed out by Cai et al. (1999), a direct comparison of transformation efficiency rates for papaya is difficult because the method used to measure efficiency differ among reports. The transformation efficiency, as described here, can be compared to those reported by Fitch et al. (1990, 1992) and Cheng et al. (1996). Our 2.7% efficiency was higher than the former (0.42%) but lower than the latter (15.9%). Cabrera-Ponce et al. (1995) and Cai et al. (1999) obtained 100% efficiency, while Mahon et al. (1996) obtained 41% efficiency, based on the percentage of bombarded plates that produced transgenic plants. Under these terms, our efficiency was 44%.
Tennant et al. (1994) showed that PRSV isolates from different geographic regions can overcome the resistance of transgenic papayas. A differing spectrum of resistance was seen among the transgenic papayas obtained in this study. Some R0 plants had a very narrow spectrum, being resistant only to the homologous isolate; while others were resistant to more than one isolate tested. All double-resistant plants were resistant to the Brazilian and to the Hawaiian isolate. None of the R0 line was shown to be resistant to PRSV BR and PRSV TH, and susceptible to PRSV HA. None of the R0 line was resistant to PRSV HA and PRSV TH, and susceptible to PRSV BR. When R1 and R2 populations were challenged with different Brazilian isolates, such as PRSV BA-CA and PRSV DF (Lima et al., 2002), resistant plants were seen in all of them, regardless of whether they descended from a single, double or even triple-resistant R0 plant. These results, together with the fact that the degree of similarity of the cp genes among the Brazilian isolates of PRSV is very high (Lima et al., 2002), strongly indicate that a variety with a broad spectrum of resistance to PRSV isolates from Brazil can be developed out of the 13 R1 and R2 populations selected.
A transgenic plant containing a transcriptionally active PRSV cp transgene is not necessarily resistant to the homologous PRSV isolate, suggesting that something more than an active cp gene is necessary to get resistance. Tennant et al. (2001) were the first to demonstrate RNA-mediated protection in transgenic papaya expressing the cp gene from PRSV. The authors suggested that a posttranscriptional gene silencing (PTGS) mechanism would be responsible for the protection seen in these transgenic plants. The R0, R1 and R2 plants obtained in this study, which have an untranslatable cp gene and are resistant to PRSV, are further evidence that suggest a RNA-mediated nature for this protection. Further analysis of these lines expressing the cp gene from PRSV BR is necessary to define which molecular mechanism is responsible for the resistance observed here.
ACKNOWLEDGMENT
Dr. Richard Manshardt (University of Hawaii) and Dr. F. Zee (USDA Repository, Hawaii) for kindly providing the three different batches of papaya fruits used in this study. Dr. Maria José A. Sampaio and Dr. Orlando S. Passos, from Embrapa, for their help in the establishing the collaborative work between Embrapa and Cornell University, which made this work possible.
LITERATURE CITED
Accepted for publication on 06/06/2005
Corresponding Author: Manoel Teixeira Souza Júnior
- AN, G. Development of plant promoter expression vectors and their use for analysis of differential activity of nopaline synthase promoter in transformed tobacco cells. Plant Physiology 81: 86-91. 1986.
- BARBOSA, F.R. & PAGUIO, O.R. Vírus da mancha anelar do mamoeiro: Incidência e efeito na produção do mamoeiro (Carica papaya L.). Fitopatologia Brasileira 7:365-373. 1982.
- CABANES-BASTOS, E., DAY, A. & LICHTENSTEIN, C. A sensitive and simple assay for neomycin phosphotransferase II activity in transgenic tissue. Gene 77:169-176. 1989.
- CABRERA-PONCE, J., VEGAS-GARCIA, A. & HERRERA-ESTRELLA, L. Herbicide resistant transgenic papaya plants produced by an efficient particle bombardment transformation method. Plant Cell Reports 15: 1-7. 1995.
- CAI, W., GONSALVES, C., TENNANT, P., FERMIN, G., SOUZA JR., M.T., SARINDU, N., JAN, F.J., ZHU, H.Y. & GONSALVES, D.A protocol for efficient transformation and regeneration of Carica papaya L. In Vitro Cellular & Development Biology - Plant 35:61-69. 1999.
- CHENG, Y.H., YANG, J.S. & YEH, S.D. Efficient transformation of papaya by coat protein gene of papaya ringspot virus mediated by Agrobacterium following liqui-phase wounding of embryogenic tissues with caborundum. Plant Cell Reports 16:127-132. 1996.
- COSTA, A.S., CARVALHO, A.M. DE & KAMADA, S. Constatado o mosaico do mamoeiro em São Paulo. O Agronômico 21:38-43. 1969.
- FEINBERG, A. & VOGELSTEIN, B. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Analytical Biochem 132:6-13. 1983.
- FITCH, M. High frequency somatic embryogenesis and plant regeneration from papaya hypocotyl callus. Plant Cell Tissue Organ Culture 32:205-212. 1993.
- FITCH, M., MANSHARDT, R., GONSALVES, D., SLIGHTOM, J. & SANFORD, J. Stable transformation of papaya via microprojectile bombardment. Plant Cell Reports 9:189-194. 1990.
- FITCH, M., MANSHARDT, R., GONSALVES, D., SLIGHTOM, J. & SANFORD, J. Virus resistant papaya plants derived from tissues bombarded with the coat protein gene of papaya ringspot virus. Bio/Technology 10:1466-1472. 1992.
- GONSALVES, D. Control of papaya ringspot virus in papaya: A case study. Annu. Rev. Phytopathol. 36:415-437. 1998.
- GONSALVES, D. & ISHII, I. Purification and serology of papaya ringspot virus. Phytopathology 70:1028-32. 1980.
- JAN, F.J. Roles of nontarget DNA and viral gene length in influencing multi-virus resistance through homology-dependent gene silencing. Ph.D. Dissertation. Cornell University. 1998.
- LIMA, R.C.A., SOUZA JR., M.T., RIBEIRO, G.P. & LIMA, J.A.A. Sequences of the coat protein gene from Brazilian isolates of Papaya ringspot virus Fitopatologia Brasileira 27:174-180. 2002.
- MAHON, R.E., BATESON, M.F., CHAMBERLAIN, D.A., HIGGINS, C.M., DREW, R.A. & DALE, J.L. Transformation of an Australian variety of Carica papaya using microprojectile bombardment. Australian Journal of Plant Physiology 23:679-685. 1996.
- MANSHARDT, R.M. Papaya. In: Biotechnology of Perennial Fruit Crops." Hammerschlag, F.A. & Litz, R.E. (Eds.), CAB International, Wallingford, England, UK. 1992. pp.489-511.
- MANSHARDT, R.M. 'UH Rainbow' papaya. University Hawaii College Tropical Agric. Hum. Resour. Germplasm G-1. 2p. 1998.
- FILHO, P.E.M., SOUZA JÚNIOR, M.T., DANTAS, J.L.L., NICKEL, O. & GONSALVES, D. Avaliação da resistência de mamoeiros transgênicos a um isolado do Papaya ringspot virus Ftopatologia Brasileira 29:146-146. 2004. (Resumo)
- MURASHIGE, T. & SKOOG, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum 15:473-497. 1962.
- NAGAO, M.A. & FURUTANI, S.C. Improving germination of papaya seed by density separation, potassium nitrate, and gibberellic acid. HortScience 21:1439-1440. 1986.
- NAPOLI, C., LEMIEUX, C. & JORGENSEN, R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2:279-289. 1990.
- PURCIFULL, D.E., EDWARDSON, J.R., HIEBERT, E. & GONSALVES, D. In: Viruses of plants: Descriptions and lists from the VIDE database. Brunt, A.A., Crabtree, K., Dallwitz, M.J., Gibbs, A.J. & Watson, L. (Eds.), CAB International, Wallingford, England, UK. 1996. pp.874-877.
- QUEMADA, H., L'HOSTIS, B., GONSALVES, D., REARDON, I.M., HEINRIKSON, R., HIEBERT, E.L., SIEU, L.C. & SLIGHTOM, J.L. The nucleotide sequences of the 3'-terminal regions of papaya ringspot virus strains W and P. Journal of General Virology 71:203-210. 1990.
- SAMBROOK, J., FRITSCH, E. & MANIATIS, T. Molecular Cloning: a Laboratory Manual. Cold Spring Harbour Laboratory. NY. 1991.
- SANFORD, J.C. & JOHNSON, S.A. The concept of parasite-derived resistance: deriving resistance genes from the parasite own genome. Journal of Theoretical Biology 115:395-405. 1985.
- SANFORD, J.C., SMITH, F.D. & RUSSELL, J.A. Optimizing the biolistic process for different biological applications. In: Wu, R. (Ed.) Methods in Enzymology; Recombinant DNA, Part H., Academis Press, Inc. 1993. pp.483-509.
- SOUZA JR., M.T. Analysis of the resistance in genetically engineered papaya against papaya ringspot potyvirus, partial characterization of the PRSV.Brazil.Bahia isolate, and development of transgenic papaya for Brazil (Ph.D. Dissertation) Cornell University. 1999.
- TENNANT, P.F., GONSALVES, C., LING, K.S., FITCH, M.M.M., MANSHARDT, R., SLIGHTOM, J.L. & GONSALVES, D. Differential protection against papaya ringspot virus isolates in coat protein gene transgenic papaya and classically cross-protected papaya. Phytopathology 84:1359-1366. 1994.
- TENNANT, P.F., FERMIN, G., FITCH, M.M., MANSHARDT, R.M., SLIGHTOM, J.L. & GONSALVES, D. Papaya ringspot virus resistance of transgenic Rainbow and SunUp is affected by gene dosage, plant development, and coat protein homology. European Journal of Plant Pathology 107:645-653. 2001.
- YEH, S.D. & GONSALVES, D. Evaluation of induced mutants of papaya ringspot virus for control by cross protection. Phytopathology 74:1086-1091. 1984.
- YEH, S.D., JAN, F.J., CHIANG, C.H., DOONG, T.J., CHEN, M.C., CHUNG, P.H. & BAU, H.J. Complete nucleotide sequence and genetic organization of papaya ringspot virus RNA. Journal of General Virology 73:2531-2541. 1992.
Publication Dates
-
Publication in this collection
17 Oct 2005 -
Date of issue
Aug 2005
History
-
Received
06 June 2005