ABSTRACT
The macaw palm, Acrocomia aculeata (Jacq.) Lodd. ex Mart., is a Brazilian native species with great potential for biofuel production. The aim of this work was to analyze macaw palm genetic diversity to structure and assist in the definition of sampling strategies for germplasm banks. Forty-six microsatellite markers, from which seven polymorphic markers were used to evaluate 103 macaw palm individuals collected from different Brazilian locations. The polymorphic markers were used to generate a dissimilarity matrix by weighted index. The imaging of genetic variability was realized by 3D projection of matrix dissimilarity. Sixty-seven individuals had their ITS region sequenced and aligned, and the mutations found were used to generate a haplotype network. The average genetic distance identified between individuals was 76.2%, ranging from 3.7 to 100%. Genetic variability structure was not found. ITS region sequencing of the 67 individuals revealed four polymorphic sites, defining four haplotypes. The results of this study suggest that historically, macaw populations were strongly connected, indicating a recent population expansion of the species. The results indicate that macaw genetic variety sampling should focus on effective collection in selected locations. Areas such as Caatinga and Humid Chaco however, could present new sources of genetic variability, and should be studied.
Keywords:
Microsatellites; ITS region; Biofuel
RESUMO
A macaúba, Acrocomia aculeata (Jacq.) Lodd. ex Mart.), é uma das espécies nativas brasileiras com maior potencial para a produção de biocombustíveis. O objetivo deste trabalho foi analisar a diversidade e estruturação genética de macaúba para auxiliar na definição de estratégias de amostragem para bancos de germoplasma. Quarenta e seis marcadores microssatélites, dos quais sete marcadores polimórficos foram utilizados para avaliar 103 indivíduos de macaúba coletados em diferentes regiões do Brasil. Os marcadores polimórficos foram utilizados para gerar uma matriz de dissimilaridade pelo índice ponderado. A visualização da variabilidade genética foi realizada através de uma projeção 3D da matriz de dissimilaridade. Sessenta e sete destes indivíduos tiveram sua região ITS sequenciada e alinhada, e as mutações encontradas foram utilizadas para gerar uma rede de haplótipos. A distância genética média identificada entre os indivíduos foi de 76,2%, variando de 3,7 a 100%. Não foi encontrada estruturação da variabilidade genética. O sequenciamento da região ITS dos 67 indivíduos revelou quatro sítios polimórficos, definindo quatro haplótipos. Os resultados obtidos neste estudo sugerem que as populações de macaúba estiveram fortemente conectadas historicamente, indicando recente expansão populacional da espécie. Os resultados indicam que a amostragem da variabilidade genética de macaúba deve estar focada em coletas mais robustas em poucos locais, mas áreas como Caatinga e Chaco Úmido podem apresentar novas fontes de variabilidade genética, e devem ser melhor estudadas.
Palavras-Chave:
Microssatélites; Região ITS; Biodiesel
INTRODUCTION
The macaw palm (Acrocomia Acrocomia aculeata (Jacq.) Lodd. ex Mart.: Arecaceae) is an arborescent, tropical, perennial and typically Brazilian palm. It presents a wide geographical distribution, being found from Paraná State in Brazil to Central America and the south of the United States. This palm tree has a single straight trunk (stipe) that can reach 20 meters, and presents a high incidence of dark thorns, especially in the node region (characteristic of the genera). The species is diploid (2n = 30) and flowers in August to November, emitting yellow flowers in inflorescences with pronounced protogyny (Scariot et al., 1991Scariot AO, Lleras E, Hay JD. Reproductive biology of the palm Acrocomia aculeata in Central Brazil. Biotropica. 1991;23(1):12-22.; Montoya et al., 2015Montoya SG, Motoike SY, Kuki KN, Oliveira CM, Honorio ICG. Registro da presença e danos causados por coleópteros em macaúba. Pesqui Florest Bras. 2015;35(82):159-62.). Despite favoring cross-fertilization, the macaw palm is considered to have a mixed mating system (Nucci et al., 2008Nucci SM, Azevedo-Filho JA, Colombo CA, Priolli RH, Coelho RM, Mata TL, et al. Development and characterization of microsatellites markers from the macaw. Mol Ecol Resour. 2008;8(1):224-6. PMid:21585762.; Mengistu et al., 2016Mengistu FG, Motoike SY, Cruz CD. Molecular characterization and genetic diversity of the macaw palm ex situ germplasm collection revealed by microsatellite markers. Diversity. 2016;8(4):20.). The species has areas of open vegetation with high solar incidence as its most common habitat, and it is recognized as a soil fertility indicator. Its role as a colonizer of deforested areas is also significant, being commonly found in pasture areas (Scariot et al., 1991Scariot AO, Lleras E, Hay JD. Reproductive biology of the palm Acrocomia aculeata in Central Brazil. Biotropica. 1991;23(1):12-22.).
The macaw palm can be used in many ways, being directly connected to familial communities and traditional customs. Fruits are the most useful portion of the plant, and mesocarp is consumed in natura or to make candies and jellies. Mesocarp oil is used to produce soap, lubricants, cosmetics, liqueurs, ice creams and biofuels (Lorenzi, 2006Lorenzi GMAC. Acrocomia aculeata (Jacq.) Lodd. ex Mart. - Arecaceae: bases para o extrativismo sustentével [thesis]. Curitiba: Universidade Federal do Paraná; 2006.; Vianna et al., 2017Vianna SA, Berton LHC, Pott A, Guerreiro SMC, Colombo CA. Biometric Characterization of Fruits and Morphoanatomy of the Mesocarp of Acrocomia Species (Arecaceae). Int J Biol. 2017;9(3):78-92.). The charcoal produced by carbonization of the endocarp is considered of high quality to produce raw materials when compared to eucalyptus charcoal (Silva et al., 1986Silva JC, Barrichelo LEG, Brito JO. Endocarpos de babaçu e macaúba comparados à madeira de Eucalyptus grandis para produção de carvão vegetal. IPEF. 1986;34:31-4.; Boas et al., 2010Boas MAV, Carneiro ADCO, Vital BR, Carvalho AMML, Martins MA. Efeito da temperatura de carbonização e dos resíduos de macaúba na produção de carvão vegetal. Sci For. 2010;38(87):481-90.). The kernel is a nobble compound and can be consumed in natura or in the form of “paçoca”, and its oil has high nutritive value, being compared to olive oil (Hiane et al., 2005Hiane PA, Ramos Filho MM, Ramos MIL, Macedo MLR. Óleo da polpa e amêndoa de bocaiúva, Acrocomia aculeata (jacq.) lodd. Caracterização e composição em ácidos graxos. Braz J Food Technol. 2005;8(3):256-9.; Lanes et al., 2015Lanes CM, Motoike SY, Kuki KN, Nick C, Freitas RD. Molecular characterization and population structure of the macaw palm, Acrocomia aculeata (Arecaceae), ex situ germplasm collection using microsatellites markers. J Hered. 2015;106(1):102-12. PMid:25425677.). Other related uses of the macaw palm are palm heart extraction from apical meristem, fabrication of masonry, stakes and walls, and production of sap wine (Lima et al., 2003Lima ES, Felfili JM, Marimon BS, Scariot A. Diversidade, estrutura e distribuição espacial de palmeiras em um cerrado sensu stricto no Brasil Central. Rev Bras Bot Braz J Bot. 2003;26(3):361-70.; Oliveira et al., 2015Oliveira DA, Moreira PA, Melo AF Jr, Pimenta MAS. Potencial da biodiversidade vegetal da Região Norte do Estado de Minas Gerais. Unimontes Científica. 2015;8(1):23-34.).
An important characteristic of the macaw palm is its distribution in low rainfall areas, being common in Cerrado, mainly in Goiás, Mato Grosso, Mato Grosso do Sul, Minas Gerais and São Paulo States. Furthermore, many studies have highlighted its fitness for consortium systems. In its initial growth stages, intercropping with small size crops like soybean and common bean is recommended. After about two years, the palms are bigger and the population is better established, being able to combine with different food and energy crops such as corn and sunflower. When the palms reach the reproductive stage, crop-livestock consortium can be employed, a cultivation system that has almost zero carbon balance of the chain (Bhering, 2010Bhering LL. O potencial da macaúba como oleífera para biocombustíveis. Biodieselbr. 2010;3(18):60-1.).
In 2002, the Brazilian government launched the Brazilian Program for Biofuels to develop technologies for biodiesel production. This program has the aim of reducing fossil fuel consumption, promoting the use of renewable energy sources and reducing greenhouse gases. Brazilian Federal Law number 13263/2016 established that from March 2017, it is mandatory to mix 8% biodiesel with diesel oil in Brazil, evidencing the strategic importance given to it by the Brazilian government (BRASIL, 2016Brasil. Lei nº 13.263, de 23 de março de 2016. Diário Oficial da União, Brasília, 24 de março de 2016.). The macaw palm has stood out as one of the most promising species for biodiesel and biokerosene production. The pulp oil presents high levels of oleic acid and reduced levels of polyunsaturated fatty acids, important features for oil stability (Hiane et al., 2005Hiane PA, Ramos Filho MM, Ramos MIL, Macedo MLR. Óleo da polpa e amêndoa de bocaiúva, Acrocomia aculeata (jacq.) lodd. Caracterização e composição em ácidos graxos. Braz J Food Technol. 2005;8(3):256-9.; Aguieiras et al., 2014Aguieiras EC, Cavalcanti-Oliveira ED, Castro AM, Langone MA, Freire DM. Biodiesel production from Acrocomia aculeata acid oil by (enzyme/ enzyme) hydroesterification process: use of vegetable lipase and fermented solid as low-cost biocatalysts. Fuel. 2014;135:315-21.). It is important to mention that the macaw palm presents potential to produce about 4,500 liters of biodiesel per hectare (Lanes et al., 2015Lanes CM, Motoike SY, Kuki KN, Nick C, Freitas RD. Molecular characterization and population structure of the macaw palm, Acrocomia aculeata (Arecaceae), ex situ germplasm collection using microsatellites markers. J Hered. 2015;106(1):102-12. PMid:25425677.). By comparison, soybean, the main source for biodiesel production in Brazil, has an average yield below 700 liters per hectare (Companhia Nacional de Abastecimento, 2017Companhia Nacional de Abastecimento. Monitoramento agrícola: safra 2016/17. Acomp. Safra Bras. Grãos. 2017;4:1-161.).
The exploitation of macaw palm is historically carried out through extraction. Genetic improvement is one of the pillars of domestication and intensive exploitation of crops. The understanding of genetic variability of the species and its relationship with the environment is of great importance to provide a basis for successfully breeding programs, in order to choose the best methods and locations for sampling as well as the existence of lineages adapted to different environmental conditions, helping to select crosses and develop enhanced cultivars.
Given the potential of the macaw palm as a source of vegetable oil for the biodiesel industry as well as its many uses, this study aimed to investigate variability and genetic structure of the macaw palm to assist in the definition of strategies for genetic variability sampling for the construction of germplasm banks and breeding programs.
2. MATERIALS AND METHODS
2.1. Genetic material
In this study, we used 64 macaw palm trees sampled from different regions of Minas Gerais and São Paulo States (MAC01 to MAC64), and 39 accesses from Active Germplasm Bank of Macauba (BAG-Macauba) at Universidade Federal de Viçosa (UFV-Brazil), located at Araponga-MG-Brazil (BAG01 to BAG39), were used for genetic diversity analysis. Geographic positions of each palm tree were determined with a GPS device (except BAG01, BAG02 and BAG04 accesses, which were not georeferenced). Leaf samples were collected, stored on ice and sent to the laboratory of Bioquímica Genética de Plantas of the Instituto de Biotecnologia Aplicada à Agropecuária (BIOAGRO) at the Universidade Federal de Viçosa, where the samples were stored at -80 ºC.
2.2. DNA Extraction
Genomic DNA was extracted following the CTAB method with modifications (Doyle and Doyle, 1990Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13-5.). About 10 mg of leaf tissue was lyophilized and powdered, homogenized in extraction buffer [1.4 mol.L-1 ofNaCl; 20 mmol.L-1 of EDTA; 100 mmol.L-1 of Tris pH 8.0; 2% CTAB (v.v-1); 2% PVP (m.v-1); 0.2% β-mercaptoetanol (v.v-1)], shaken and incubated at 65 ºC for 1 hour. Then, the organic phase was removed in a solution of chloroform/ isoamyl alcohol (24:1). The DNA was precipitated in cold isopropanol, washed in ethanol 70%, incubated in TE containing RNAse at 37 ºC and stored at -20 ºC.
2.3. Amplification and microsatellite marker analysis
Thirty-six microsatellite markers developed by Nucci et al. (2008)Nucci SM, Azevedo-Filho JA, Colombo CA, Priolli RH, Coelho RM, Mata TL, et al. Development and characterization of microsatellites markers from the macaw. Mol Ecol Resour. 2008;8(1):224-6. PMid:21585762. were used in the genetic analysis. Due the small number of polymorphic loci described for the species, five microsatellite markers described for Astrocaryum sciophilum by Girod et al. (2009)Girod C, Tollon-Cordet C, Pfunder M, Sarrazin E, Samadi S, Boisselier-Dubayle MC, et al. Characterization of 11 microsatellites in the palm Astrocaryum sciophilum (Arecaceae). Mol Ecol Resour. 2009;10:1098-105. and five microsatellite markers described for Bactris gasipaes by Rodrigues et al. (2004)Rodrigues DP, Vinson C, Ciampi AY, Farias IP, Lemes MR, Astolfi-Filho S, et al. Novel microsatellite markers for Bactris gasipaes (Palmae). Mol Ecol Notes. 2004;4:575-6. were added to the study. The amplification reactions were carried out in 15 µ.L containing 30 ng of genomic DNA, 12 mmol.L-1 of Tris-HCl pH 8.3; 60 mmol.L-1 of KCl; 1.5-4 mmol.L-1 of MgCl2; 100 µmol.L- 1 of each deoxynucleotide; 0-2% of formamide (v.v-1); 0.33 µmol.L-1 of each primer; and 1 U of Taq DNA Polymerase (Phoneutria), using the following conditions: 94 ºC for 4 minutes; 35 cycles started at 94 ºC for 40 seconds, the specific annealing temperature for each primer for 40 seconds and 72 ºC for 1 minute; and a final step at 72 ºC for 7 minutes. The amplification products were submitted to electrophoresis on 10% polyacrylamide gel in TAE buffer at 130 V for about 4 hours. The products were visualized using a silver nitrate protocol as described by Qu et al. (2005)Qu L, Li X, Wu G, Yang N. Efficient and sensitive method of DNA silver staining in polyacrylamide gels. Electrophoresis. 2005;26(1):99-101. PMid:15624131. with modifications. The fragment length was confirmed using a 50 bp marker (Fermentas).
The dissimilarity matrix was constructed using the complement of weighted similarity index (Cruz et al., 2011Cruz CD, Ferreira FM, Pessoni LA. Biometria aplicada ao estudo da diversidade genética. Visconde do Rio Branco: Suprema; 2011.). The Mantel test (MANTEL 1967Mantel N. Detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27(2):209-20. PMid:6018555.) was performed to verify the presence of a correlation between the dissimilarity matrix and the geographic distance matrix. Cluster analysis was performed using 3D projection (Cruz et al., 2011Cruz CD, Ferreira FM, Pessoni LA. Biometria aplicada ao estudo da diversidade genética. Visconde do Rio Branco: Suprema; 2011.).
2.4. Amplification, sequencing and ITS region analysis
The internal transcribed ribosomal 18S-26S spacer (ITS region) was sequenced in 67 individuals (MAC01 to MAC13, MAC15 to MAC60, BAG01 to BAG08) using universal primers ITS4 and ITS5 (White et al., 1990White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols. 1990;18(1):315-22.). The amplification reaction was performed in a total volume of 20 µL containing 60 ng of genomic DNA, 1X Go-Taq Buffer, 3.75 mmol.L-1 of MgCl2, 250 µmol.L-1 of each deoxynucleotide, 0.5 µmol.L-1 of each primer, 1.25 µL of DMSO and 1.25 U of Go-Taq DNA Polymerase (Promega), using the following cycling parameters: initial denaturation at 72 ºC for 45 seconds; 35 cycles started at 94 ºC for 1 minute, 62 ºC for 1 minute and 72 ºC for 45 seconds, and a final step at 72 ºC for 5 minutes. The amplification products were visualized on 1.2 agarose gel, purified with PCR Exosap (Affymetrix) and sequenced. The generated sequences were edited and aligned using Sequencher software version 4.1.4 (Gene Codes Corporation). A haplotype network was constructed using Network software version 4.6.1.1 (Fluxus Technology Ltd) through Median Joining method (Bandelt et al., 1999Bandelt HJ, Forster P, Rohl A. Median-Joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16(1):37-48. PMid:10331250.).
3. RESULTS
3.1. Microsatellite reactions
Among the 46 microsatellite markers used in this study, only seven (Table 1) were reproducible and polymorphic between an initial sample of genotypes: five markers described for A. aculeata (Nucci et al., 2008Nucci SM, Azevedo-Filho JA, Colombo CA, Priolli RH, Coelho RM, Mata TL, et al. Development and characterization of microsatellites markers from the macaw. Mol Ecol Resour. 2008;8(1):224-6. PMid:21585762.), one marker described for A. sciophilum (Girod et al., 2009Girod C, Tollon-Cordet C, Pfunder M, Sarrazin E, Samadi S, Boisselier-Dubayle MC, et al. Characterization of 11 microsatellites in the palm Astrocaryum sciophilum (Arecaceae). Mol Ecol Resour. 2009;10:1098-105.) and one marker described for B. gasipaes (Rodrigues et al., 2004Rodrigues DP, Vinson C, Ciampi AY, Farias IP, Lemes MR, Astolfi-Filho S, et al. Novel microsatellite markers for Bactris gasipaes (Palmae). Mol Ecol Notes. 2004;4:575-6.). The number of alleles ranged from four (Aacu07) to nine (Aacu12), with an average of 6.7 alleles per locus.
Sequences (F: forward, R: reverse) of seven microsatellite loci analyzed in 103 Acrocomia aculeata individuals, showing annealing temperature (Tm), bands amplitude, number of alleles and Genbank accession number.
Tabela 1
Sequências (F: forward, R: reverse) de sete locos microssatélites analisados em 103 indivíduos de Acrocomia aculeata, mostrando temperatura de anelamento (Tm), amplitude das bandas, número de alelos e número de acesso no Genbank.
3.2. Evaluation of genetic diversity by microsatellite markers
The average genetic distance from the dissimilarity matrix generated by the complement of weighted similarity indexes was 76.2%. The MAC05 and MAC06 individuals were identified as the most similar (93.6% similarity), while several pairs of individuals presented 100% dissimilarity (data not shown). The Mantel test showed a significant correlation between genetic distance and geographic distance matrices (r = 0.34; P = 0 for 10,000 permutations).
Cluster analysis by 3D projection method (Figure 1) showed a correlation of r = 0.52 with the original dissimilarity matrix. In this 3D projection, information about haplotype of the individuals (see item 3.3) was added to check for the presence of structuring between genetic diversity from microsatellite markers and from ITS region haplotypes.
3D projection grouping of genetic dissimilarity matrix between 103 Acrocomia aculeata individuals, and haplotypes generated by sequencing ITS region of 67 individuals (see Figures 2 and 3).
Figura 1
Agrupamento por projeção 3D da matriz de dissimilaridade genética entre 103 indivíduos de Acrocomia aculeata, e haplótipos gerados pelo sequenciamento da região ITS de 67 indivíduos (ver Figuras 2 e 3).
3.3. Evaluation of genetic diversity by ITS region
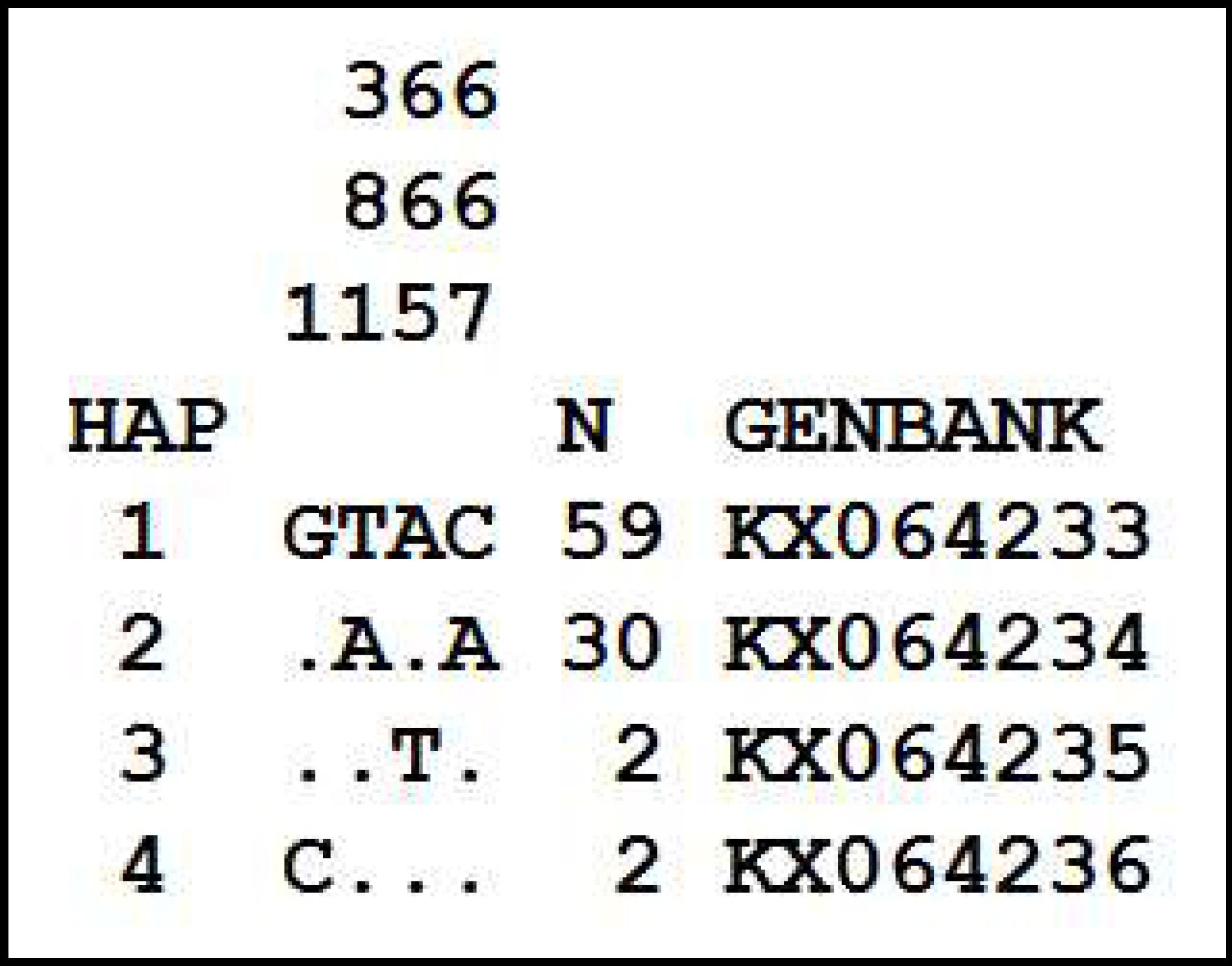
The sequencing of the ITS region of 67 individuals generated a 667 bp fragment that presents four polymorphic sites (three base transversions and one base inversion), of which two showed ambiguity (intraindividual polymorphism). Evaluation of these two sites revealed only two parental haplotypes, indicating that the individuals which showed intraindividual polymorphism present both 1 and 2 haplotypes. A total of four haplotypes were generated, differing by at most two mutations. Figure 2 shows information about the four haplotypes, and their geographic distribution is shown in Figure 3.
Sequence alignment of variable sites in the ITS region, defining four ITS haplotypes of Acrocomia aculeata. Each fragment spans 667 bases. Dots indicate similarity to haplotype 1. Numbers on the top indicate the nucleotide position. The number of occurrences of each haplotype in 67 individuals (N) and the accession number in Genbank are shown.
Figura 2
Alinhamento de sequências de sítios variáveis na região ITS, definindo quatro haplótipos ITS de Acrocomia aculeata. Cada fragmento se estende por 667 bases. Pontos indicam similaridade ao haplótipo 1. Números no topo indicam a posição do nucleotídeo. O número de ocorrências de cada haplótipo nos 67 indivíduos (N) e o número de acesso no Genbank são mostrados.
Brazilian States map showing the geographic distribution of four haplotypes generated by sequencing the ITS region of 67 Acrocomia aculeata individuals. (A) Overview of the accesses distribution. (B) Accesses collected in Pará (PA), Paraíba (PB) and Pernambuco (PE) states. (C) Accesses collected in Mato Grosso do Sul (MS), Minas Gerais (MG) and São Paulo (SP) states. The semi filled circle symbol indicates individuals that presente intraindividual polymorphism (combining haplotypes 1 and 2).
Figura 3
Mapa de estados brasileiros mostrando a distribuição geográfica de quatro haplótipos gerados pelo sequenciamento da região ITS de 67 indivíduos de Acrocomia aculeata. (A) Visão geral da distribuição dos acessos. (B) Acessos coletados nos estados do Pará (PA), Paraíba (PB) e Pernambuco (PE). (C) Acessos coletados nos estados de Mato Grosso do Sul (MS), Minas Gerais (MG) e São Paulo (SP). O símbolo círculo semipreenchido indica indivíduos que apresentaram polimorfismo intraindividual (combinação dos haplótipos 1 e 2).
4. DISCUSSION
The average number of alleles found (6.7) agrees with other studies involving the macaw palm. Nucci et al. (2008)Nucci SM, Azevedo-Filho JA, Colombo CA, Priolli RH, Coelho RM, Mata TL, et al. Development and characterization of microsatellites markers from the macaw. Mol Ecol Resour. 2008;8(1):224-6. PMid:21585762. found an average of five alleles per locus in a study using eight microsatellite loci to analyze forty-three individuals from natural A. aculeata populations from Minas Gerais and São Paulo States. Lanes et al. (2015)Lanes CM, Motoike SY, Kuki KN, Nick C, Freitas RD. Molecular characterization and population structure of the macaw palm, Acrocomia aculeata (Arecaceae), ex situ germplasm collection using microsatellites markers. J Hered. 2015;106(1):102-12. PMid:25425677. found an average of nine alleles per locus using six microsatellite loci in a study with accesses of a germplasm bank with samples collected from different regions of Brazil.
Transferability of microsatellite markers between different species is an alternative in the search for new sources of genetic variability, mainly in less studied species. While Barbará et al. (2007)Barbará T, Palma-Silva C, Paggi GM, Bered F, Fay MF, Lexer C. Cross species transfer of nuclear microsatellite markers: potential and limitations. Mol Ecol. 2007;16(18):3759-67. PMid:17850543. considered low transferability of microsatellite markers between monocot genera, other works showed good results in studies with genetic diversity in palm trees (Mistura et al., 2012Mistura CC, Barbieri RL, Castro CM, Priori D, Villela JCB. Transferibilidade de marcadores microssatélites de coco (Cocos nucifera) para butiá (Butia odorata). Magistra. 2012;24:360-9.; Ramos et al., 2012Ramos SLF, Macêdo JLV, Lopes MTG, Batista JS, Formiga KM, da Silva PP, et al. Microsatellite loci for tucumã of Amazonas (Astrocaryum aculeatum) and amplification in other Arecaceae. Am J Bot. 2012;99(12):508-10. PMid:23211567.). In this study, Asci01 and BACg152 loci (described for A. sciophilum and B. gasipaes, respectively) showed allele numbers consistent with the overall mean (seven and six loci, respectively; Table 1), indicating that loci transferability between palm genera can provide more genetic data for the macaw palm.
Clustering analysis using 3D projection was performed using the data generated by microsatellite markers. The generated projection presented a correlation of r = 0.52 with the original dissimilarity matrix, a value below the ideal level. In the same way, a dendrogram generated by UPGMA method (data not shown) showed regular setting with the genetic dissimilarity matrix. In these cases, an increase in the number of polymorphic loci was recommended. Thus, transferability of molecular markers from genera related to A. aculeata, as well as employing other methods to generate genetic data, like non-specific markers (AFLP, RAPD) and development of new microsatellite markers, must be considered for future studies.
The 3D graph projection allows viewing the presence of genetic diversity according to the region sampled. However, even though the Mantel test showed a significant correlation between genetic dissimilarity and geographic distance matrices (r = 0.34), it was not possible to visualize a consistent clustering that indicates structuring of genetic variability according to the geographic distribution. In the same way, haplotype distribution shown in the 3D projection (Figure 1) did not show apparent structuring with microsatellite genetic diversity.
Analysis of ITS sequences of 67 palm trees revealed only four mutation sites, leading to the formation of four haplotypes (Figure 2), a low level of polymorphism when compared to similar studies, such as Oliveira et al. (2010)Oliveira LO, Rossi AAB, Martins ER, Batista FRC, Silva RS. Molecular phylogeography of Carapichea ipecacuanha, an amphitropical shrub that occurs in the understory of both semideciduous and evergreen forests. Mol Ecol. 2010;19(7):1410-22. PMid:20298468. and Khan et al. (2014)Khan G, Zhang F, Gao Q, Fu PC, Zing R, Wang J, et al. Molecular phylogeography and intraspecific divergence of Spiraea alpina (Rosaceae) distributed in the Qinghai-Tibetan Plateau and adjacent regions inferred from nrDNA. Biochem Syst Ecol. 2014;57:278-86.. However, studies of the ITS region in other species from the Arecaceae family were not found in the literature, making inferences about this low level of polymorphism difficult. Two of the four haplotypes occurred in regions of distinct biomes which are poorly sampled (Figure 3). Haplotype 3 was found in two palm trees from Humid Chaco (BAG03 and BAG08). This biome is a mosaic of ecosystems combining forests with savanna and presents high rainfall and reduced seasonality, characteristics that distinguish it from other biomes where the macaw palm is found, mainly in Atlantic Forest and Cerrado (World Wild Fund, 2017aWorld Wild Fund. Southeastern South America, in Argentina, Paraguay, and Brazil. Gland: WWF; 2017a [cited 2017 Jul 06]. Available from: http://www.worldwildlife.org/ecoregions/nt0708
http://www.worldwildlife.org/ecoregions/...
). Haplotype 4 was found in two palm trees from Caatinga biome (BAG01 and BAG02), a warm climate and low precipitation region that presents six to eleven months of drought per year (World Wild Fund, 2017bWorld Wild Fund. Northern South America: Northeastern Brazil. Gland: WWF; 2017b [cited 2017 Jul 06]. Available from: http://www.worldwildlife.org/ecoregions/nt1304
http://www.worldwildlife.org/ecoregions/...
). The peculiarities of these two biomes indicates a possible existence of macaw palm lineages with particular characteristics, which has implications both in conservation and genetic variability sampling for germplasm banks, aiming at the genetic improvement of the species. Therefore, more detailed studies of genetic variability in these regions are needed.
The low level of polymorphism found in the ITS region (Figure 2), coupled with the geographic distribution of the accesses (Figure 3), suggests agreement with category V of phylogeographic patterns of Avise et al. (1987)Avise JC, Arnold J, Ball RM, Bermingham E, Lamb T, Neigel JE, et al. Intraspecific phylogeography: the mitochondrial dna bridge between population genetics and systematics. Annu Rev Ecol Syst. 1987;18:489-522.. In this category, common lineages are widely found in all regions, while related lineages are confined to limited regions. The results in this study suggest that macaw palm populations were strongly connected in the past, indicating a recent population expansion of the species (Avise et al., 1987). Similar results have been found in studies with tree species in Brazil (Novaes et al., 2010Novaes RML, Lemes JP Fo, Ribeiro RA, Lovato MB. Phylogeography of Plathymenia reticulata (Leguminosae) reveals patterns of recent range expansion towards northeastern Brazil and southern Cerrados in Eastern Tropical South America. Mol Ecol. 2010;19(5):985-98. PMid:20149092., 2013Novaes RML, Riveiro RA, Lemes-Filho JP, Lovato MB. Concordance between phylogeographical and biogeographical patterns in the Brazilian Cerrado: diversification of the endemic tree Dalbergia miscolobium (Fabaceae). PLoS One. 2013;8(12):e82198. PMid:24312640.). Moreover, recent population expansion of typical Cerrado species was suggested in a climate modeling study (Bonaccorso et al., 2006Bonaccorso E, Koch I, Peterson AT. Pleistocene fragmentation of Amazon species’ ranges. Divers Distrib. 2006;12:157-64.).
According to the results found in this study, the sampling of genetic variability of the macaw palm should focus on detailed sampling of a few locations rather than sampling a large number of regions. However, more studies should be conducted to better understand the genetic variability of the macaw palm in Caatinga and Humid Chaco biomes, as well as in other regions where the occurrence of Acrocomia genera is reported, which can lead to new strategies for genetic variability sampling for the species.
5.CONCLUSION
Acrocomia aculeata presents genetic diversity, but this diversity apparently shows no geographic structuring, probably due to some recent population expansion process in the species. Thus, sampling strategies for genetic variability should focus on more detailed sampling of a few locations. However, areas like Caatinga and Humid Chaco can provide new sources of genetic variability, and should be better investigated in future studies.
6. ACKNOWLEDGMENTS
The authors would like to thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and the Fundação de Amparo à Pesquisa do Estado de Minas Gerais (Fapemig) for their financial support, and the group of professor Sérgio Yoshimitsu Motoike for germplasm contribution. LCCS received a master’s scholarship grant from the CNPq, and RCL received a scholarship grant from the CNPq. Special thanks to professor Maurilio Alves Moreira, for his life of dedication to Brazilian research and higher education.
7. REFERENCES
- Aguieiras EC, Cavalcanti-Oliveira ED, Castro AM, Langone MA, Freire DM. Biodiesel production from Acrocomia aculeata acid oil by (enzyme/ enzyme) hydroesterification process: use of vegetable lipase and fermented solid as low-cost biocatalysts. Fuel. 2014;135:315-21.
- Avise JC, Arnold J, Ball RM, Bermingham E, Lamb T, Neigel JE, et al. Intraspecific phylogeography: the mitochondrial dna bridge between population genetics and systematics. Annu Rev Ecol Syst. 1987;18:489-522.
- Bandelt HJ, Forster P, Rohl A. Median-Joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16(1):37-48. PMid:10331250.
- Barbará T, Palma-Silva C, Paggi GM, Bered F, Fay MF, Lexer C. Cross species transfer of nuclear microsatellite markers: potential and limitations. Mol Ecol. 2007;16(18):3759-67. PMid:17850543.
- Bhering LL. O potencial da macaúba como oleífera para biocombustíveis. Biodieselbr. 2010;3(18):60-1.
- Boas MAV, Carneiro ADCO, Vital BR, Carvalho AMML, Martins MA. Efeito da temperatura de carbonização e dos resíduos de macaúba na produção de carvão vegetal. Sci For. 2010;38(87):481-90.
- Bonaccorso E, Koch I, Peterson AT. Pleistocene fragmentation of Amazon species’ ranges. Divers Distrib. 2006;12:157-64.
- Brasil. Lei nº 13.263, de 23 de março de 2016. Diário Oficial da União, Brasília, 24 de março de 2016.
- Companhia Nacional de Abastecimento. Monitoramento agrícola: safra 2016/17. Acomp. Safra Bras. Grãos. 2017;4:1-161.
- Cruz CD, Ferreira FM, Pessoni LA. Biometria aplicada ao estudo da diversidade genética. Visconde do Rio Branco: Suprema; 2011.
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13-5.
- Girod C, Tollon-Cordet C, Pfunder M, Sarrazin E, Samadi S, Boisselier-Dubayle MC, et al. Characterization of 11 microsatellites in the palm Astrocaryum sciophilum (Arecaceae). Mol Ecol Resour. 2009;10:1098-105.
- Hiane PA, Ramos Filho MM, Ramos MIL, Macedo MLR. Óleo da polpa e amêndoa de bocaiúva, Acrocomia aculeata (jacq.) lodd. Caracterização e composição em ácidos graxos. Braz J Food Technol. 2005;8(3):256-9.
- Khan G, Zhang F, Gao Q, Fu PC, Zing R, Wang J, et al. Molecular phylogeography and intraspecific divergence of Spiraea alpina (Rosaceae) distributed in the Qinghai-Tibetan Plateau and adjacent regions inferred from nrDNA. Biochem Syst Ecol. 2014;57:278-86.
- Lanes CM, Motoike SY, Kuki KN, Nick C, Freitas RD. Molecular characterization and population structure of the macaw palm, Acrocomia aculeata (Arecaceae), ex situ germplasm collection using microsatellites markers. J Hered. 2015;106(1):102-12. PMid:25425677.
- Lima ES, Felfili JM, Marimon BS, Scariot A. Diversidade, estrutura e distribuição espacial de palmeiras em um cerrado sensu stricto no Brasil Central. Rev Bras Bot Braz J Bot. 2003;26(3):361-70.
- Lorenzi GMAC. Acrocomia aculeata (Jacq.) Lodd. ex Mart. - Arecaceae: bases para o extrativismo sustentével [thesis]. Curitiba: Universidade Federal do Paraná; 2006.
- Mantel N. Detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27(2):209-20. PMid:6018555.
- Mengistu FG, Motoike SY, Cruz CD. Molecular characterization and genetic diversity of the macaw palm ex situ germplasm collection revealed by microsatellite markers. Diversity. 2016;8(4):20.
- Mistura CC, Barbieri RL, Castro CM, Priori D, Villela JCB. Transferibilidade de marcadores microssatélites de coco (Cocos nucifera) para butiá (Butia odorata) Magistra. 2012;24:360-9.
- Montoya SG, Motoike SY, Kuki KN, Oliveira CM, Honorio ICG. Registro da presença e danos causados por coleópteros em macaúba. Pesqui Florest Bras. 2015;35(82):159-62.
- Novaes RML, Lemes JP Fo, Ribeiro RA, Lovato MB. Phylogeography of Plathymenia reticulata (Leguminosae) reveals patterns of recent range expansion towards northeastern Brazil and southern Cerrados in Eastern Tropical South America. Mol Ecol. 2010;19(5):985-98. PMid:20149092.
- Novaes RML, Riveiro RA, Lemes-Filho JP, Lovato MB. Concordance between phylogeographical and biogeographical patterns in the Brazilian Cerrado: diversification of the endemic tree Dalbergia miscolobium (Fabaceae). PLoS One. 2013;8(12):e82198. PMid:24312640.
- Nucci SM, Azevedo-Filho JA, Colombo CA, Priolli RH, Coelho RM, Mata TL, et al. Development and characterization of microsatellites markers from the macaw. Mol Ecol Resour. 2008;8(1):224-6. PMid:21585762.
- Oliveira DA, Moreira PA, Melo AF Jr, Pimenta MAS. Potencial da biodiversidade vegetal da Região Norte do Estado de Minas Gerais. Unimontes Científica. 2015;8(1):23-34.
- Oliveira LO, Rossi AAB, Martins ER, Batista FRC, Silva RS. Molecular phylogeography of Carapichea ipecacuanha, an amphitropical shrub that occurs in the understory of both semideciduous and evergreen forests. Mol Ecol. 2010;19(7):1410-22. PMid:20298468.
- Qu L, Li X, Wu G, Yang N. Efficient and sensitive method of DNA silver staining in polyacrylamide gels. Electrophoresis. 2005;26(1):99-101. PMid:15624131.
- Ramos SLF, Macêdo JLV, Lopes MTG, Batista JS, Formiga KM, da Silva PP, et al. Microsatellite loci for tucumã of Amazonas (Astrocaryum aculeatum) and amplification in other Arecaceae. Am J Bot. 2012;99(12):508-10. PMid:23211567.
- Rodrigues DP, Vinson C, Ciampi AY, Farias IP, Lemes MR, Astolfi-Filho S, et al. Novel microsatellite markers for Bactris gasipaes (Palmae). Mol Ecol Notes. 2004;4:575-6.
- Scariot AO, Lleras E, Hay JD. Reproductive biology of the palm Acrocomia aculeata in Central Brazil. Biotropica. 1991;23(1):12-22.
- Silva JC, Barrichelo LEG, Brito JO. Endocarpos de babaçu e macaúba comparados à madeira de Eucalyptus grandis para produção de carvão vegetal. IPEF. 1986;34:31-4.
- Vianna SA, Berton LHC, Pott A, Guerreiro SMC, Colombo CA. Biometric Characterization of Fruits and Morphoanatomy of the Mesocarp of Acrocomia Species (Arecaceae). Int J Biol. 2017;9(3):78-92.
- White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols. 1990;18(1):315-22.
- World Wild Fund. Southeastern South America, in Argentina, Paraguay, and Brazil. Gland: WWF; 2017a [cited 2017 Jul 06]. Available from: http://www.worldwildlife.org/ecoregions/nt0708
» http://www.worldwildlife.org/ecoregions/nt0708 - World Wild Fund. Northern South America: Northeastern Brazil. Gland: WWF; 2017b [cited 2017 Jul 06]. Available from: http://www.worldwildlife.org/ecoregions/nt1304
» http://www.worldwildlife.org/ecoregions/nt1304
Publication Dates
-
Publication in this collection
2017
History
-
Received
27 May 2016 -
Accepted
22 Aug 2017