Abstracts
The episodes of diarrhea caused by neonatal bovine rotavirus group A (BoRVA) constitute one of the major health problems in the calf rearing worldwide. The main G (VP7) and P (VP4) genotypes of BoRVA strains involved in the etiology of diarrhea in calves are G6P[1], G10P[11], G6P[5], and G8P[1]. However, less frequently, other G and P genotypes have been described in BoRVA strains identified in diarrheic fecal samples of calves. This study describes the identification and molecular characterization of an emerging genotype (G6P[11]) in BoRVA strains involved in the etiology of a diarrhea outbreak in beef calves in a cattle herd of high production in extensive management system. The diarrhea outbreak, which showed high morbidity (60%) and lethality (7%) rates, occurred in calves (n= 384) Nelore (Bos indicus) up to 30-day-old from the State of Mato Grosso do Sul, Brazil. BoRVA was identified in 80% (16/20) of the fecal samples analyzed by polyacrylamide gel electrophoresis (PAGE) technique. In all PAGE-positive fecal samples were amplified products with 1,062-bp and 876-bp in the RT-PCR assays for VP7 (G type) and VP4 (VP8*) (P type) of BoRVA, respectively. The nucleotide sequence analysis of VP7 and VP4 genes of four wild-type BoRVA strains showed G6-III P[11]-III genotype/lineage. The G6P[11] genotype has been described in RVA strains of human and animal hosts, however, in calves this genotype was only identified in some cross-sectional studies and not as a single cause of diarrhea outbreaks in calves with high morbidity and lethality rates as described in this study. The monitoring of the G and P genotypes of BoRVA strains involved in diarrhea outbreaks in calves is important for both animal and public health by allowing the identification of the most frequent genotypes, the characterization of novel genotypes and to identify reassortments with genotypes described in animal and human hosts. The results of this study show the importance of the monitoring of the genotypes of BoRVA strains involved in episodes of bovine neonatal diarrhea as for characterization of frequency of occurrence and pathogenic potential of uncommon genotypes as for monitoring of the emergency of different BoRVA genotypes not included in commercial vaccines.
Calf; enteric infection; rotaviruses; VP7 gene; VP4 gene
Os episódios de diarreia neonatal ocasionados pelo rotavírus bovino grupo A (BoRVA) constituem-se em um dos principais problemas sanitários na criação de bezerros em todo o mundo. Os principais genotipos G (VP7) e P (VP4) de cepas de BoRVA envolvidos na etiologia da diarreia em bezerros são G6P[1], G10P[11], G6P[5] e G8P[1]. No entanto, com menor frequência, outros genotipos G e P têm sido descritos em cepas de BoRVA identificadas em amostras de fezes diarreicas de bezerros. Este estudo descreve a identificação e caracterização molecular de um genotipo emergente (G6P[11]) em cepas de BoRVA envolvidas na etiologia de um surto de diarreia em bezerros de um rebanho bovino de corte de alta produção em sistema de manejo extensivo. O surto, que apresentou altas taxas de morbidade (60%) e de letalidade (7%), ocorreu em bezerros (n=384) da raça Nelore (Bos indicus) com até 30 dias de idade, provenientes do estado do Mato Grosso do Sul, Brasil. O BoRVA foi identificado em 80% (16/20) das amostras fecais analisadas pela técnica de eletroforese em gel de poliacrilamida (PAGE). Em todas as amostras fecais PAGE-positivas foi possível a amplificação por RT-PCR de produtos com 1.062 pb e 876 pb referentes aos genes VP7 (G tipo) e VP4 (VP8*) (P tipo), respectivamente, de BoRVA. A análise da sequência de nucleotídeos dos genes VP7 e VP4 de quatro cepas de BoRVA demonstrou a presença do genotipo/linhagem G6-III P[11]-III. O genotipo G6P[11] tem sido descrito em cepas de RVA de hospedeiros humanos e animais. Contudo, em bezerros, este genotipo foi apenas identificado em alguns estudos transversais e não como a única causa de surtos de diarreia em bezerros com altas taxas de morbidade e letalidade como descrito neste estudo. O monitoramento dos genotipos G e P de cepas de BoRVA envolvidos em surtos de diarreia em bezerros é relevante tanto para a saúde animal quanto para a saúde pública por possibilitar a identificação dos genotipos mais frequentes, a caracterização de novos genotipos e por identificar reassortments com genotipos descritos em hospedeiros humanos e animais. Os resultados deste estudo demonstram a importância do monitoramento dos genotipos de cepas de BoRVA envolvidas em surtos de diarreia neonatal bovina tanto para a caracterização da frequência de ocorrência e potencial patogênico de genotipos incomuns quanto para o monitoriamento da emergência de genotipos distintos daqueles incluídos em vacinas comerciais.
Bezerro; infecção entérica; rotavirose; gene VP7; gene VP4
LIVESTOCK DISEASES ANIMAIS DE PRODUÇÃO
Severe diarrhea outbreak in beef calves (Bos indicus) caused by G6P[11], an emergent genotype of bovine rotavirus group A
Surto de diarreia neonatal em bezerros de corte (Bos indicus) ocasionado por um genotipo emergente G6P[11] de rotavírus bovino grupo A
Thais N.S. Medeiros; Elis Lorenzetti; Alice F. Alfieri; Amauri A. Alfieri* * Corresponding author: alfieri@uel.br
Laboratory of Animal Virology, Department of Veterinary Preventive Medicine, Universidade Estadual de Londrina (UEL), Rodovia Celso Garcia Cid, Campus Universitário, Cx. Postal 10.011, Londrina, PR 86057-970, Brazil
ABSTRACT
The episodes of diarrhea caused by neonatal bovine rotavirus group A (BoRVA) constitute one of the major health problems in the calf rearing worldwide. The main G (VP7) and P (VP4) genotypes of BoRVA strains involved in the etiology of diarrhea in calves are G6P[1], G10P[11], G6P[5], and G8P[1]. However, less frequently, other G and P genotypes have been described in BoRVA strains identified in diarrheic fecal samples of calves. This study describes the identification and molecular characterization of an emerging genotype (G6P[11]) in BoRVA strains involved in the etiology of a diarrhea outbreak in beef calves in a cattle herd of high production in extensive management system. The diarrhea outbreak, which showed high morbidity (60%) and lethality (7%) rates, occurred in calves (n= 384) Nelore (Bos indicus) up to 30-day-old from the State of Mato Grosso do Sul, Brazil. BoRVA was identified in 80% (16/20) of the fecal samples analyzed by polyacrylamide gel electrophoresis (PAGE) technique. In all PAGE-positive fecal samples were amplified products with 1,062-bp and 876-bp in the RT-PCR assays for VP7 (G type) and VP4 (VP8*) (P type) of BoRVA, respectively. The nucleotide sequence analysis of VP7 and VP4 genes of four wild-type BoRVA strains showed G6-III P[11]-III genotype/lineage. The G6P[11] genotype has been described in RVA strains of human and animal hosts, however, in calves this genotype was only identified in some cross-sectional studies and not as a single cause of diarrhea outbreaks in calves with high morbidity and lethality rates as described in this study. The monitoring of the G and P genotypes of BoRVA strains involved in diarrhea outbreaks in calves is important for both animal and public health by allowing the identification of the most frequent genotypes, the characterization of novel genotypes and to identify reassortments with genotypes described in animal and human hosts. The results of this study show the importance of the monitoring of the genotypes of BoRVA strains involved in episodes of bovine neonatal diarrhea as for characterization of frequency of occurrence and pathogenic potential of uncommon genotypes as for monitoring of the emergency of different BoRVA genotypes not included in commercial vaccines.
Index terms: Calf, enteric infection, rotaviruses, VP7 gene, VP4 gene.
RESUMO
Os episódios de diarreia neonatal ocasionados pelo rotavírus bovino grupo A (BoRVA) constituem-se em um dos principais problemas sanitários na criação de bezerros em todo o mundo. Os principais genotipos G (VP7) e P (VP4) de cepas de BoRVA envolvidos na etiologia da diarreia em bezerros são G6P[1], G10P[11], G6P[5] e G8P[1]. No entanto, com menor frequência, outros genotipos G e P têm sido descritos em cepas de BoRVA identificadas em amostras de fezes diarreicas de bezerros. Este estudo descreve a identificação e caracterização molecular de um genotipo emergente (G6P[11]) em cepas de BoRVA envolvidas na etiologia de um surto de diarreia em bezerros de um rebanho bovino de corte de alta produção em sistema de manejo extensivo. O surto, que apresentou altas taxas de morbidade (60%) e de letalidade (7%), ocorreu em bezerros (n=384) da raça Nelore (Bos indicus) com até 30 dias de idade, provenientes do estado do Mato Grosso do Sul, Brasil. O BoRVA foi identificado em 80% (16/20) das amostras fecais analisadas pela técnica de eletroforese em gel de poliacrilamida (PAGE). Em todas as amostras fecais PAGE-positivas foi possível a amplificação por RT-PCR de produtos com 1.062 pb e 876 pb referentes aos genes VP7 (G tipo) e VP4 (VP8*) (P tipo), respectivamente, de BoRVA. A análise da sequência de nucleotídeos dos genes VP7 e VP4 de quatro cepas de BoRVA demonstrou a presença do genotipo/linhagem G6-III P[11]-III. O genotipo G6P[11] tem sido descrito em cepas de RVA de hospedeiros humanos e animais. Contudo, em bezerros, este genotipo foi apenas identificado em alguns estudos transversais e não como a única causa de surtos de diarreia em bezerros com altas taxas de morbidade e letalidade como descrito neste estudo. O monitoramento dos genotipos G e P de cepas de BoRVA envolvidos em surtos de diarreia em bezerros é relevante tanto para a saúde animal quanto para a saúde pública por possibilitar a identificação dos genotipos mais frequentes, a caracterização de novos genotipos e por identificar reassortments com genotipos descritos em hospedeiros humanos e animais. Os resultados deste estudo demonstram a importância do monitoramento dos genotipos de cepas de BoRVA envolvidas em surtos de diarreia neonatal bovina tanto para a caracterização da frequência de ocorrência e potencial patogênico de genotipos incomuns quanto para o monitoriamento da emergência de genotipos distintos daqueles incluídos em vacinas comerciais.
Termos de indexação: Bezerro, infecção entérica, rotavirose, gene VP7, gene VP4.
INTRODUCTION
Neonatal diarrhea is one of the most important diseases of calves worldwide, causing economic losses to both dairy and beef cattle herds (Holland 1990). Diarrhea in calves is a multi-factorial disease associated with predisposing and determinant factors (environment, management, nutrition, immunology, and microorganisms). There are many microorganisms involved in neonatal calf diarrhea such as bacteria (Escherichia coli K99 enteropathogenic), protozoans (Cryptosporidium sp. and Eimeria sp.), and viruses (rotavirus and coronavirus) (Oliveira Filho et al. 2007, Stipp et al. 2009, Izzo et al. 2011, Blanchard 2012).
Bovine group A rotavirus (BoRVA) is one the most important viral agents of neonatal diarrhea in calves worldwide (Barreiros et al. 2004, Alfieri et al. 2006, Badaracco et al. 2012, Midgley et al. 2012). The rotavirus belongs to the Reoviridae family and Rotavirus genus, and is composed of a triple-layered protein capsid. The genome is formed by 11 double-stranded RNA (dsRNA) segments (Estes & Kapikian 2007). Due to the VP6 protein, which composes the middle layer capsid, the rotavirus (RV) is classified into eight distinct serogroups (A-H) (Matthijnssens et al. 2012).
The outer layer VP7 and VP4 proteins of rotavirus group A (RVA) strains determine the binary viral classification in G (glycoprotein) and P (protease-sensitive) genotypes, respectively (Estes & Kapikian 2007). Currently, 27 G (VP7) and 37 P (VP4) genotypes of RVA have been described in human and animal hosts (Matthijnssens et al. 2011, Trojnar et al. 2013). The most common combination of G and P genotypes of BoRVA strains isolated from diarrheic calves are G6P[1] (NCDV-Lincoln strain), G10P[11] (B223 strain), G6P[5] (UK strain), and G8P[1] (A5 strain) (Fukai et al. 1999, Gulati et al. 1999, Alfieri et al. 2004, Barreiros et al. 2004). However, with lower frequency other G and P genotypes are sporadically detected in calves and buffalos worldwide such as G6P[3], G10P[3], G8P[7], G6P[7], and G6P[11] (Martella et al. 2003, Manuja et al. 2008, Park et al. 2011).
The G6P[11] is an emerging RVA genotype that was previously described in diarrheic fecal samples from animals (cattle, lamb, and pig) and humans worldwide (Munford et al. 2007, Martini et al. 2008, Monini et al. 2008, De Grazia et al. 2011, Sherif et al. 2011, Yamamoto et al. 2011, Gazal et al. 2012, Midgley et al. 2012). In Brazilian cattle herds the BoRVA genotype G6P[11] was only identified in diarrheic fecal samples in lower frequency in cross-sectional epidemiological studies (Brito et al. 2000, Alfieri et al. 2004, Barreiros et al. 2004, Buzinaro et al. 2009, Caruzo et al. 2010, Silva et al. 2012). However, there are no reports of the G6P[11] genotype involvement as a single causative agent of diarrheal outbreak in Brazilian beef cattle herds.
The G6 genotype has been divided into five (I-V) distinct lineages. The lineages G6-I, contain human rotavirus (RV) strains, and the G6-IV contain bovine RV strains. The G6-II, G6-III, and G6-V lineages are composed by both human and animal RV strains (Badaracco et al. 2013). The P[11] genotype has been classified in three (I-III) lineages. The lineages P[11]-I and P[11]-II are composed by human RV strains, while the lineage P[11]-III are composed by human and bovine RV strains (Badaracco et al. 2013). This report describes the identification of an emergent BoRVA G6P[11] genotype as cause of a severe neonatal diarrhea outbreak in a beef cattle herd.
MATERIALS AND METHODS
The herd was composed by 2,970 Nelore (Bos indicus) cows reared extensively. The reproductive management included a breeding season of 90 days. The diarrhea outbreak, with duration of 20 to 25 days affected 384 calves that were up to 30 days of age, resulting in elevated morbidity (60%) and lethality (7%) rates. All diarrheic calves were medicated with wide spectrum antibiotics, but response to treatment was inconsistent and 27 calves died. Diarrheic fecal samples (n=20) were collected diagnosis during September 2011, and were maintained at -20°C until used.
All diarrheic fecal samples were submitted to nucleic acid extraction using a combination of phenol/chloroform/isoamyl alcohol (25:24:1) and silica/guanidinium isothiocyanate methods described by Alfieri et al. (2006). The cell culture adapted BoRVA NCDV strain (G6P[1]) and aliquots of buffer Tris-Ca2+ were included in all viral RNA extraction and RT-PCR procedures as positive and negative controls, respectively.
The presence of dsRNA of RVA in the diarrheic fecal samples was evaluated using the 7.5% polyacrylamide gel electrophoresis (PAGE) technique (Pereira et al. 1983) followed by silver staining according to Herring et al. (1982). The fecal samples were submitted to RT-PCR assays using previously described consensus primers for the amplification of VP7 (G type) and VP4/VP8*(P type) of RVA (Gouvea et al. 1990, 1993, Gentsch et al. 1992, Martella et al. 2006).
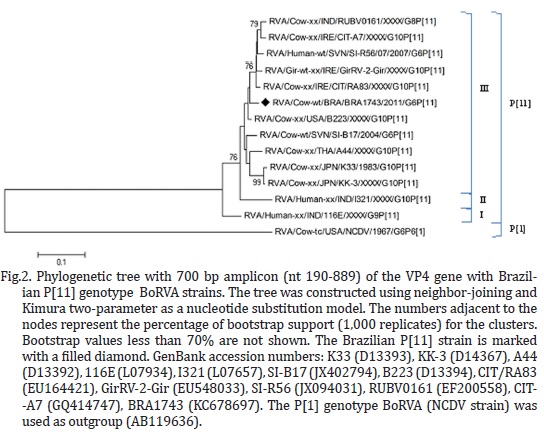
Four samples (BRA1743, BRA1744, BRA1752, and BRA1758) were selected for nucleotide sequence analysis based on the amount of fecal sample, intensity of dsRNA bands in PAGE, and the RT-PCR results. RT-PCR products of VP7 and VP4 genes were purified using the GFX PCR DNA and Gel Band purification kit (GE Healthcare, Little Chalfont, Buckinghamshire, UK) and quantified with a Qubit® Fluorometer (Invitrogen Life Technologies, Eugene, OR, USA). The DNA sequences were obtained using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) with the ABI3500 Genetic Analyzer sequencer (Applied Biosystems, Foster City, CA, USA) using the forward and reverse primers. Sequence quality analyses were performed using Phred and CAP3 software and the sequences were accepted if the base quality was ≥ 20. Multiple and pairwise alignments were realized in MEGA version 5.05 with CLUSTAL W (version 1.4) and the nucleotide identity matrix was constructed using BioEdit software version 7.0.8.0. Phylogenetic trees based on the nucleotide sequences obtained were created using the neighbor-joining method with the Kimura two-parameter model using MEGA 5.05 software. The bootstrap probabilities of each node were calculated with 1,000 replications.
RESULTS
BoRVA was detected in 80% (16 out 20) of diarrheic fecal samples analyzed by PAGE technique. All samples showed long electrophoretic profile with the 4:2:3:2 distribution patterns of the 11 dsRNA segments. In the RT-PCR assay, the same 16 PAGE-positive fecal samples amplified 1,062 bp and 876 bp products of VP7 and VP4 genes, respectively.
A 100% of nucleotide (nt) identity was demonstrated between the VP7 and VP4 nt sequences of four wild-type Brazilian BoRVA strains (BRA1743, BRA1744, BRA1752, and BRA1758); these sequences are available in GenBank with the accession numbers KC678693 to KC678700. During the VP7 nt sequence analysis, the four Brazilian wild-type BoRVA strains demonstrated elevated nt identity (95.2 to 96.4%) with human (Hun-3) and bovine (B102) strains, that belong to the G6-III lineage (Fig.1). The VP4 nt sequence analysis of the BoRVA field strains showed high nt identity (95.8 to 96.8%) with GirRV2 (G6P[11]) and B223 (G10P[11]) BoRVA strains, that belong to P[11]-III lineage (Fig.2).
DISCUSSION
The high rate (80%) of BoRVA positive diarrheic fecal samples identified by PAGE technique and RT-PCR assay demonstrated that the neonatal diarrhea outbreak was caused by this virus. Additionally, all fecal samples were still negative for bovine coronavirus by semi-nested-PCR (Takiuchi et al. 2006) and for Cryptosporidium spp. by modified Ziehl-Neelsen technique (Henriksen & Pohlenz 1981) (data not shown). Eimeriosis is reported more frequently in calves older than two months of life and the diarrheic calves were refractory to the first generation wide spectrum antibiotic therapy, so the diagnosis of Eimeria sp and enteropathogenic bacteria were not included in the present study.
Most studies relating to neonatal calf diarrhea have been cross-sectional investigation with the frequency of BoRVA detection varying from 9.9% (31/331) to 30% (435/1462) (Barreiros et al. 2004, Caruzo et al. 2010, Badaraco et al. 2012). In a longitudinal study, the BoRVA was detected in 11% (11/100) of fecal samples analyzed (Oliveira Filho et al. 2007). Brito et al. (2000) described an outbreak of neonatal diarrhea in a dairy cattle herd from midwest Brazil where the G10P[11] (B223-like strain), a common genotype of BoRVA was found in 68.75% (11/16) of the fecal samples evaluated.
The P[11] genotype is frequently related to BoRVA strains in association with the G10 genotype (B223 prototype strain) (Beg et al. 2010). The G6P[11] is not the most commonly combination described in cattle herds worldwide. However, this emergent BoRVA genotype has already been diagnosed in surveys done in calves from Europe and Argentina, and sheep in India (Monini et al. 2008, Gazal et al. 2012, Midgley et al. 2012, Badaracco et al. 2013). Moreover, the G6P[11] genotype has been identified in RVA strains from human hosts from Africa (Sherif et al. 2011), Asia (Yamamoto et al. 2011), Europe (De Grazia et al. 2011, Steyer et al. 2013), Oceania (Chandrahasen et al. 2010), and America (Brazil) (Munford et al. 2007, Martini et al. 2008). Moreover, cattle can be a reservoir of rotavirus G6P[11] genotype for human hosts (Martella et al. 2010).
The BoRVA G6P[11] genotype was previously described in Brazilian cattle herds. Some cross-sectional epidemiological surveys for the determination of the most frequent BoRVA genotypes done in stored fecal samples have identified the combination G6P[11] in detection rates that varied from 2% (1/34) to 34.3% (12/35) of the samples analyzed (Alfieri et al. 2004, Barreiros et al. 2004, Buzinaro et al. 2009, Caruzo et al. 2010, Silva et al. 2012). However, this is the first report of a severe neonatal diarrhea outbreak caused by a single infection of the G6P[11] genotype in Nelore calves (Bos indicus). Additionally, the BoRVA genotype G6P[11] identified in this diarrheal outbreak belongs to the G6-III lineage that contains human and bovine RV strains, distant from the most usual G6-IV lineage of bovine RVA strains that includes the G6P[5] (UK strain) and G6P[1] (NCDV-Lincoln strain) prototypes.
Neonatal diarrhea is the main important disease in calf worldwide (Holland 1990). However, two characteristics that predominate in Brazilian beef cattle herds make the diarrhea outbreaks are not so frequent. The extensive management system used within the country decreases the transmission risk of enteropathogenic microorganisms and Nelore (Bos indicus) cattle, the most prevalent breed in the country, is characterized by rusticity when compared with Bos taurus or crossbreed calves.
Some aspect that might increase the risk of neonatal diarrhea outbreak is the reproductive management adopted throughout Brazilian beef cattle herds with a breeding season of 90-120 days. The concentration of birth increases the number of susceptible animals favoring the transmission of enteric pathogens. In this situation, calf sanitary management should include risk analysis of neonatal diarrhea and the adoption of measures for control and prophylaxis of the infections. Therefore, two conditions of sanitary management may have contributed to the severity, morbidity, and mortality of neonatal diarrhea outbreak observed in this beef cattle herd. First, the large number of susceptible animals concentrated in a calving area, which is an impacting risk factor. Second, the non-adoption of a neonatal diarrhea immunoprophylaxis programs with cow vaccination. These two conditions (management x immunity) predispose the occurrence of neonatal diarrhea that in association with the presence of this emergent strain of BoRVA for which the herd immunity was probably low or inexistence, contributed to the severity of the outbreak.
Around the world, the monitoring of the G and P genotypes of RVA strains involved in diarrhea outbreaks is important for animal and public health. The identification of a new G and/or P genotypes or the uncommon combination of G and P types in reassortment strains has great impact on the effectiveness of vaccination programs against rotavirus implemented in both human and livestock animals such as cattle and pigs. The G6P[11] is an uncommon combination of the G and P genotypes in BoRVA strains and their detection as single genotype involved in a severe diarrhea outbreak in beef calves reared extensively serve as an alert to the importance of this emergent BoRVA genotype for the health of calves. Since cattle can be a reservoir of G6P[11] strains for human hosts this study might be of public health concern. Although this survey is one of the few studies in the literature that has accompanied all the outbreak, with marked features of high morbidity and mortality in cattle infected with rotavirus G6P[11] in Brazil.
CONCLUSION
Only with the molecular characterization of BoRVA strains will be possible to determine the most frequent genotypes as well as to monitor the emergence of genotypes not included in commercial vaccines, and to evaluate the pathogenic potential of uncommon BoRVA genotypes involved in calf diarrhea outbreak. With the use of molecular tools this study allowed the first identification of an emerging genotype, G6P[11], of BoRVA involved in the etiology of a neonatal diarrhea outbreak in a Brazilian beef cattle herd.
Acknowledgements.- This study was supported by the following Brazilian institutes: National Counsel of Scientific and Technological Development (CNPq), Brazilian Federal Agency for Support and Evaluation of Graduate Education (CAPES), Financing of Studies and Projects (FINEP), and the Araucaria Foundation (FAP/PR). A.A. Alfieri and A.F. Alfieri are recipients of CNPq fellowships.
REFERENCES
Alfieri A.F., Barreiros M.A.B., Leite J.P.G., Richtzenhain L.J. & Alfieri A.A. 2004. G and P genotypes of group A rotavirus strains circulating in calves in Brazil, 1996-1999. Vet. Microbiol. 99:167-173.
Alfieri A.A., Parazzi M.E., Takiuchi E., Médici K.C. & Alfieri A.F. 2006. Frequency of group A rotavirus in diarrhoeic calves in Brazilian cattle herds, 1998-2002. Trop. Anim. Health Prod. 38:521-526.
Badaracco A., Garaicoechea L., Rodríguez D., Uriarte E.L., Odeón A., Bilbao G., Galarza R., Abdala A., Fernandez F. & Parreño V. 2012. Bovine rotavirus strains circulating in beef and dairy herds in Argentina from 2004 to 2010. Vet. Microbiol. 158:394-399.
Badaracco A., Garaicoechea L., Matthijnssens J., Uriarte E.L., Odeón A., Bilbao G., Fernandez F., Parra G.I. & Parreño V. 2013. Phylogenetic analysis of typical bovine rotavirus genotypes G6, G10, P[5] and P[11] circulating in Argentinean beef and dairy herds. Infect. Genet. Evol. 18:18-30.
Barreiros M.A.B., Alfieri A.F., Médici K.C., Leite J.P.G. & Alfieri A. A. 2004. G and P genotypes of group A rotavirus from diarrhoeic calves born to cows vaccinated against the NCDV (P[1], G6) rotavirus strain. J. Vet. Med. 51:104-109.
Beg S.A., Wani S.A., Hussain I. & Bhat M.A. 2010. Determination of G and P type diversity of group A rotaviruses in faecal samples of diarrhoeic calves in Kashmir, India. Lett. Appl. Microbiol. 51:595-599.
Blanchard P.C. 2012. Diagnostics of dairy and beef cattle diarrhea. Vet. Clin. North. Am., Food. Anim. Pract. 28:443-464.
Brito W.M.E.D., Munford V., Villaça A.M., Caruzo T.A.R. & Rácz M.L. 2000. Characterization of mixed infections with different strains of bovine rotavirus in an outbreak of diarrhea in dairy herds in Goiás, Brazil. Braz. J. Microbiol. 31:140-145.
Buzinaro M.G., Samara S.I., Pereira E.A.S., Fuentes D.B. & Oliveira M.C.S. 2009. Occurrence of genotypes G and P of group A rotavirus in calves in beef herds in the state of São Paulo, Brazil. Arqs Inst. Biológico, São Paulo, 76:99-105.
Caruzo T.A., Brito W.M., Munford V. & Rácz M.L. 2010. Molecular characterization of G and P-types bovine rotavirus strains from Goiás, Brazil: high frequency of mixed P-type infections. Mem. Inst. Oswaldo Cruz. 105:1040-1043.
Chandrahasen C., Grimwood K., Redshaw N., Rich F.J., Wood C., Stanley J. & Kirman J.R. 2010. Geographical differences in the proportion of human group A rotavirus strains within New Zealand during one epidemic season. J. Med. Virol. 82:897-902.
De Grazia S., Martella V., Rotolo V., Bonura F., Matthijnssens J., Bányai K., Ciarlet M. & Giammanco G.M. 2011. Molecular characterization of genotype G6 human rotavirus strains detected in Italy from 1986 to 2009. Infect. Genet. Evol. 11:1449-1455.
Estes M.K. & Kapikian A.Z. 2007. Rotaviruses, p.1918-1974. In: Knipe D.M., Howley P.M. (Eds), Fields Virology 5th ed. Lippincott Williams and Wilkins, Philadelphia.
Fukai K., Sakai T., Hirose M. & Itou T. 1999. Prevalence of calf diarrhea caused by bovine group A rotavirus carrying G serotype 8 specificity. Vet. Microbiol. 66:301-311.
Gazal S., Taku A.K. & Kumar B. 2012. Predominance of rotavirus genotype G6P[11] in diarrhoeic lambs. Vet. J. 193:299-300.
Gentsch J.R., Glass R.I., Woods P., Gouvea V., Gorziglia M., Flores J., Das B.K. & Bhan M.K. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373.
Gouvea V., Glass R.I., Woods P., Taniguchi K., Clark H.F., Forrester B. & Fang Z.Y. 1990. Polymerase Chain Reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282.
Gouvea V., Ramirez C., Li B., Santos N., Saif L.J., Clark H.F. & Hoshino Y. 1993. Restriction endonuclease analysis of the VP7 genes of human and animal rotaviruses. J. Clin. Microbiol. 31:917-923.
Gulati B.R., Nakagomi O., Koshimura Y., Nakagomi T. & Pandey R. 1999. Relative frequencies of G and P types among rotaviruses from Indian diarrheic cow and buffalo calves. J. Clin. Microbiol. 37:2074-2076.
Henriksen A. & Pohlenz J.F.L. 1981. Staining of Cryptosporidium by a modified Ziehl-Nielsen technique. Acta Vet. Scand. 22:594-596.
Herring A.J., Inglis N.F., Ojeh C.K., Snodgrass D.R. & Menzies J.D. 1982. Rapid diagnosis of rotavirus infection by direct detection of viral nucleic acid in silver-stained polyacrylamide gels. J. Clin. Microbiol. 16:473-477.
Holland R.E. 1990. Some infectious causes of diarrhea in young farm animals. Clin. Microbiol. Rev. 3:345-375.
Izzo M.M., Kirkland P.D., Mohler V.L., Perkins N.R., Gunn A.A. & House J.K. 2011. Prevalence of major enteric pathogens in Australian dairy calves with diarrhoea. Aust. Vet. J. 89:167-173.
Manuja B.K., Prasad M., Manuja A., Gulati B.R. & Prasad G. 2008. A novel genomic constellation (G10P[3]) of group A rotavirus detected from buffalo calves in northern India. Virus Res. 138:36-42.
Martella V., Ciarlet M., Pratelli A., Arista S., Terio V., Elia G., Cavalli A., Gentile M., Decaro N., Greco G., Cafiero M.A., Tempesta M. & Buonavoglia C. 2003. Molecular analysis of the VP7, VP4, VP6, NSP4, and NSP5/6 genes of a buffalo rotavirus strain: identification of the rare P[3] rhesus rotavirus-like VP4 gene allele. J. Clin. Microbiol. 41:5665-5675.
Martella V., Ciarlet M., Bányai K., Lorusso E., Cavalli A., Corrente M., Elia G., Arista S., Camero M., Desario C., Decaro N., Lavazza A. & Buonavoglia C. 2006. Identification of a novel VP4 genotype carried by a serotype G5 porcine rotavirus strain. Virology. 346:301-311.
Martella V., Bányai K., Matthijnssens J., Buonavoglia C. & Ciarlet M. 2010. Zoonotic aspects of rotaviruses. Vet. Microbiol. 140:246-255.
Matthijnssens J., Ciarlet M., McDonald S.M., Attoui H., Bányai K., Brister J.R., Buesa J., Esona M.D., Estes M.K., Gentsch J.R., Iturriza-Gómara M., Johne R., Kirkwood C.D., Martella V., Mertens P.P., Nakagomi O., Parreño V., Rahman M., Ruggeri F.M., Saif L.J., Santos N., Steyer A., Taniguchi K., Patton J.T., Desselberger U. & Van Ranst M. 2011. Uniformity of rotavirus strain nomenclature proposed by the rotavirus classification working group (RCWG). Arch. Virol. 156:1397-1413.
Matthijnssens J., Otto P.H., Ciarlet M., Desselberger U., Van Ranst M. & Johne R. 2012. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch. Virol. 157:1177-82.
Martini I.J., Gennari G.M., Martins S.S., Gouvêa V.S. & Gatti M.S.V. 2008. Changing distribution of human rotavirus serotypes during two epidemic outbreaks of gastroenteritis in Campinas, São Paulo, Brazil, 2003-2004: detection of G6 strains. J. Clin. Virol. 43: 244-246.
Midgley S.E., Bányai K., Buesa J., Halaihel N., Hjulsager C.K., Jakab F., Kaplon J., Larsen L.E., Monini M., Poljšak-Prijatelj M., Pothier P., Ruggeri F.M., Steyer A., Koopmans M. & Böttiger B. 2012. Diversity and zoonotic potential of rotaviruses in swine and cattle across Europe. Vet. Microbiol. 156:238-245.
Monini M., Cappuccini F., Battista P., Falcone E., Lavazza A. & Ruggeri F.M. 2008. Molecular characterization of bovine rotavirus strains circulating in northern Italy, 2003-2005. Vet. Microbiol. 129:384-389.
Munford V., Souza E.C., Caruzo T.A.R., Martinez M.B. & Racz M.L. 2007. Serological and molecular diversity of human rotavirus in São Paulo, Brazil. Braz. J. Microbiol. 38:459-466.
Oliveira-Filho J.P., Silva D.P.G., Pacheco M.D., Mascarini L.M., Ribeiro M.G., Alfieri A.A., Alfieri A.F., Stipp D.T., Barros B.J.P. & Borges A.S. 2007. Diarrhea in nelore calves reared extensively: clinical and etiological. Pesq. Vet. Bras. 27:419-424.
Park S.I., Matthijnssens J., Saif L.J., Kim H.J., Park J.G., Alfajaro M.M., Kim D.S., Son K.Y., Yang D.K., Hyun B.H., Kang M.I. & Cho K.O. 2011. Reassortment among bovine, porcine and human rotavirus strains results in G8P[7] and G6P[7] strains isolated from cattle in South Korea. Vet. Microbiol. 152:55-66.
Pereira H.G., Azeredo R.S., Leite J.P.G., Candeias J.A.N., Racz M.L., Linhares A.C., Gabbay Y.B. & Trabulsi J.R. 1983. Eletrophoretic study of the genome of human rotaviruses from Rio de Janeiro, São Paulo and Belém, Brazil. J. Hyg. 90:117-125.
Silva F.D.F., Gregori F., Gonçalves A.C.S., Samara S.I. & Buzinaro M.G. 2012. Molecular characterization of group A bovine rotavirus in southeastern and central-western Brazil, 2009-2010. Pesq. Vet. Bras. 32:237-242.
Sherif M.E., Esona M.D., Wang Y., Gentsch J.R., Jiang B., Glass R.I., Baker S.A. & Klena J.D. 2011. Detection of the first G6P[14] human rotavirus strain from a child with diarrhea in Egypt. Infect. Genet. Evol. 11:1436-1442.
Steyer A., Sagadin M., Kolenc M. & Poljšak-Prijatelj M. 2013. Whole genome sequence analysis of bovine G6P[11] rotavirus strain found in a child with gastroenteritis. Infect. Genet. Evol. 13:89-95.
Stipp D.T., Barry A.F., Alfieri A.F., Takiuchi E., Amude A.M. & Alfieri A.A. 2009. Frequency of BCoV detection by a semi-nested PCR assay in faeces of calves from Brazilian cattle herds. Trop. Anim. Health Prod. 41:1563-1567.
Takiuchi E., Stipp D.T., Alfieri A.F. & Alfieri A.A. 2006. Improved detection of bovine coronavirus N gene in faeces of calves infected naturally by a semi-nested PCR assay and an internal control. J. Virol. Methods 131:148-154.
Trojnar E., Sachsenroder J., Twardziok S., Reetz J., Otto P.H. & Johne R. 2013. Identification of an avian group A rotavirus containing a novel VP4 gene of close relationship to those of mammalian rotaviruses. J. Gen. Virol. 94:136-142.
Yamamoto D., Kawaguchiya M., Ghosh S., Ichikawa M., Numazaki K. & Kobayashi N. 2011. Detection and full genomic analysis of G6P[9] human rotavirus in Japan. Virus Genes 43:215-223.
Received on March 14, 2014.
Accepted for publication on May 5, 2014.
- Alfieri A.F., Barreiros M.A.B., Leite J.P.G., Richtzenhain L.J. & Alfieri A.A. 2004. G and P genotypes of group A rotavirus strains circulating in calves in Brazil, 1996-1999. Vet. Microbiol. 99:167-173.
- Alfieri A.A., Parazzi M.E., Takiuchi E., Médici K.C. & Alfieri A.F. 2006. Frequency of group A rotavirus in diarrhoeic calves in Brazilian cattle herds, 1998-2002. Trop. Anim. Health Prod. 38:521-526.
- Badaracco A., Garaicoechea L., Rodríguez D., Uriarte E.L., Odeón A., Bilbao G., Galarza R., Abdala A., Fernandez F. & Parreño V. 2012. Bovine rotavirus strains circulating in beef and dairy herds in Argentina from 2004 to 2010. Vet. Microbiol. 158:394-399.
- Badaracco A., Garaicoechea L., Matthijnssens J., Uriarte E.L., Odeón A., Bilbao G., Fernandez F., Parra G.I. & Parreño V. 2013. Phylogenetic analysis of typical bovine rotavirus genotypes G6, G10, P[5] and P[11] circulating in Argentinean beef and dairy herds. Infect. Genet. Evol. 18:18-30.
- Barreiros M.A.B., Alfieri A.F., Médici K.C., Leite J.P.G. & Alfieri A. A. 2004. G and P genotypes of group A rotavirus from diarrhoeic calves born to cows vaccinated against the NCDV (P[1], G6) rotavirus strain. J. Vet. Med. 51:104-109.
- Beg S.A., Wani S.A., Hussain I. & Bhat M.A. 2010. Determination of G and P type diversity of group A rotaviruses in faecal samples of diarrhoeic calves in Kashmir, India. Lett. Appl. Microbiol. 51:595-599.
- Blanchard P.C. 2012. Diagnostics of dairy and beef cattle diarrhea. Vet. Clin. North. Am., Food. Anim. Pract. 28:443-464.
- Brito W.M.E.D., Munford V., Villaça A.M., Caruzo T.A.R. & Rácz M.L. 2000. Characterization of mixed infections with different strains of bovine rotavirus in an outbreak of diarrhea in dairy herds in Goiás, Brazil. Braz. J. Microbiol. 31:140-145.
- Buzinaro M.G., Samara S.I., Pereira E.A.S., Fuentes D.B. & Oliveira M.C.S. 2009. Occurrence of genotypes G and P of group A rotavirus in calves in beef herds in the state of São Paulo, Brazil. Arqs Inst. Biológico, São Paulo, 76:99-105.
- Caruzo T.A., Brito W.M., Munford V. & Rácz M.L. 2010. Molecular characterization of G and P-types bovine rotavirus strains from Goiás, Brazil: high frequency of mixed P-type infections. Mem. Inst. Oswaldo Cruz. 105:1040-1043.
- Chandrahasen C., Grimwood K., Redshaw N., Rich F.J., Wood C., Stanley J. & Kirman J.R. 2010. Geographical differences in the proportion of human group A rotavirus strains within New Zealand during one epidemic season. J. Med. Virol. 82:897-902.
- De Grazia S., Martella V., Rotolo V., Bonura F., Matthijnssens J., Bányai K., Ciarlet M. & Giammanco G.M. 2011. Molecular characterization of genotype G6 human rotavirus strains detected in Italy from 1986 to 2009. Infect. Genet. Evol. 11:1449-1455.
- Estes M.K. & Kapikian A.Z. 2007. Rotaviruses, p.1918-1974. In: Knipe D.M., Howley P.M. (Eds), Fields Virology 5th ed. Lippincott Williams and Wilkins, Philadelphia.
- Fukai K., Sakai T., Hirose M. & Itou T. 1999. Prevalence of calf diarrhea caused by bovine group A rotavirus carrying G serotype 8 specificity. Vet. Microbiol. 66:301-311.
- Gazal S., Taku A.K. & Kumar B. 2012. Predominance of rotavirus genotype G6P[11] in diarrhoeic lambs. Vet. J. 193:299-300.
- Gentsch J.R., Glass R.I., Woods P., Gouvea V., Gorziglia M., Flores J., Das B.K. & Bhan M.K. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373.
- Gouvea V., Glass R.I., Woods P., Taniguchi K., Clark H.F., Forrester B. & Fang Z.Y. 1990. Polymerase Chain Reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282.
- Gouvea V., Ramirez C., Li B., Santos N., Saif L.J., Clark H.F. & Hoshino Y. 1993. Restriction endonuclease analysis of the VP7 genes of human and animal rotaviruses. J. Clin. Microbiol. 31:917-923.
- Gulati B.R., Nakagomi O., Koshimura Y., Nakagomi T. & Pandey R. 1999. Relative frequencies of G and P types among rotaviruses from Indian diarrheic cow and buffalo calves. J. Clin. Microbiol. 37:2074-2076.
- Henriksen A. & Pohlenz J.F.L. 1981. Staining of Cryptosporidium by a modified Ziehl-Nielsen technique. Acta Vet. Scand. 22:594-596.
- Herring A.J., Inglis N.F., Ojeh C.K., Snodgrass D.R. & Menzies J.D. 1982. Rapid diagnosis of rotavirus infection by direct detection of viral nucleic acid in silver-stained polyacrylamide gels. J. Clin. Microbiol. 16:473-477.
- Holland R.E. 1990. Some infectious causes of diarrhea in young farm animals. Clin. Microbiol. Rev. 3:345-375.
- Izzo M.M., Kirkland P.D., Mohler V.L., Perkins N.R., Gunn A.A. & House J.K. 2011. Prevalence of major enteric pathogens in Australian dairy calves with diarrhoea. Aust. Vet. J. 89:167-173.
- Manuja B.K., Prasad M., Manuja A., Gulati B.R. & Prasad G. 2008. A novel genomic constellation (G10P[3]) of group A rotavirus detected from buffalo calves in northern India. Virus Res. 138:36-42.
- Martella V., Ciarlet M., Pratelli A., Arista S., Terio V., Elia G., Cavalli A., Gentile M., Decaro N., Greco G., Cafiero M.A., Tempesta M. & Buonavoglia C. 2003. Molecular analysis of the VP7, VP4, VP6, NSP4, and NSP5/6 genes of a buffalo rotavirus strain: identification of the rare P[3] rhesus rotavirus-like VP4 gene allele. J. Clin. Microbiol. 41:5665-5675.
- Martella V., Ciarlet M., Bányai K., Lorusso E., Cavalli A., Corrente M., Elia G., Arista S., Camero M., Desario C., Decaro N., Lavazza A. & Buonavoglia C. 2006. Identification of a novel VP4 genotype carried by a serotype G5 porcine rotavirus strain. Virology. 346:301-311.
- Martella V., Bányai K., Matthijnssens J., Buonavoglia C. & Ciarlet M. 2010. Zoonotic aspects of rotaviruses. Vet. Microbiol. 140:246-255.
- Matthijnssens J., Ciarlet M., McDonald S.M., Attoui H., Bányai K., Brister J.R., Buesa J., Esona M.D., Estes M.K., Gentsch J.R., Iturriza-Gómara M., Johne R., Kirkwood C.D., Martella V., Mertens P.P., Nakagomi O., Parreño V., Rahman M., Ruggeri F.M., Saif L.J., Santos N., Steyer A., Taniguchi K., Patton J.T., Desselberger U. & Van Ranst M. 2011. Uniformity of rotavirus strain nomenclature proposed by the rotavirus classification working group (RCWG). Arch. Virol. 156:1397-1413.
- Matthijnssens J., Otto P.H., Ciarlet M., Desselberger U., Van Ranst M. & Johne R. 2012. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch. Virol. 157:1177-82.
- Martini I.J., Gennari G.M., Martins S.S., Gouvêa V.S. & Gatti M.S.V. 2008. Changing distribution of human rotavirus serotypes during two epidemic outbreaks of gastroenteritis in Campinas, São Paulo, Brazil, 2003-2004: detection of G6 strains. J. Clin. Virol. 43: 244-246.
- Monini M., Cappuccini F., Battista P., Falcone E., Lavazza A. & Ruggeri F.M. 2008. Molecular characterization of bovine rotavirus strains circulating in northern Italy, 2003-2005. Vet. Microbiol. 129:384-389.
- Munford V., Souza E.C., Caruzo T.A.R., Martinez M.B. & Racz M.L. 2007. Serological and molecular diversity of human rotavirus in São Paulo, Brazil. Braz. J. Microbiol. 38:459-466.
- Oliveira-Filho J.P., Silva D.P.G., Pacheco M.D., Mascarini L.M., Ribeiro M.G., Alfieri A.A., Alfieri A.F., Stipp D.T., Barros B.J.P. & Borges A.S. 2007. Diarrhea in nelore calves reared extensively: clinical and etiological. Pesq. Vet. Bras. 27:419-424.
- Park S.I., Matthijnssens J., Saif L.J., Kim H.J., Park J.G., Alfajaro M.M., Kim D.S., Son K.Y., Yang D.K., Hyun B.H., Kang M.I. & Cho K.O. 2011. Reassortment among bovine, porcine and human rotavirus strains results in G8P[7] and G6P[7] strains isolated from cattle in South Korea. Vet. Microbiol. 152:55-66.
- Pereira H.G., Azeredo R.S., Leite J.P.G., Candeias J.A.N., Racz M.L., Linhares A.C., Gabbay Y.B. & Trabulsi J.R. 1983. Eletrophoretic study of the genome of human rotaviruses from Rio de Janeiro, São Paulo and Belém, Brazil. J. Hyg. 90:117-125.
- Silva F.D.F., Gregori F., Gonçalves A.C.S., Samara S.I. & Buzinaro M.G. 2012. Molecular characterization of group A bovine rotavirus in southeastern and central-western Brazil, 2009-2010. Pesq. Vet. Bras. 32:237-242.
- Sherif M.E., Esona M.D., Wang Y., Gentsch J.R., Jiang B., Glass R.I., Baker S.A. & Klena J.D. 2011. Detection of the first G6P[14] human rotavirus strain from a child with diarrhea in Egypt. Infect. Genet. Evol. 11:1436-1442.
- Stipp D.T., Barry A.F., Alfieri A.F., Takiuchi E., Amude A.M. & Alfieri A.A. 2009. Frequency of BCoV detection by a semi-nested PCR assay in faeces of calves from Brazilian cattle herds. Trop. Anim. Health Prod. 41:1563-1567.
- Takiuchi E., Stipp D.T., Alfieri A.F. & Alfieri A.A. 2006. Improved detection of bovine coronavirus N gene in faeces of calves infected naturally by a semi-nested PCR assay and an internal control. J. Virol. Methods 131:148-154.
- Trojnar E., Sachsenroder J., Twardziok S., Reetz J., Otto P.H. & Johne R. 2013. Identification of an avian group A rotavirus containing a novel VP4 gene of close relationship to those of mammalian rotaviruses. J. Gen. Virol. 94:136-142.
- Yamamoto D., Kawaguchiya M., Ghosh S., Ichikawa M., Numazaki K. & Kobayashi N. 2011. Detection and full genomic analysis of G6P[9] human rotavirus in Japan. Virus Genes 43:215-223.
Publication Dates
-
Publication in this collection
30 Sept 2014 -
Date of issue
Aug 2014
History
-
Received
14 Mar 2004 -
Accepted
05 May 2004