Abstract
Lignin, after cellulose, is the second most abundant biopolymer on Earth, accounting for 30% of the organic carbon in the biosphere. It is considered an important evolutionary adaptation of plants during their transition from the aquatic environment to land, since it bestowed the early tracheophytes with physical support to stand upright and enabled long-distance transport of water and solutes by waterproofing the vascular tissue. Although essential for plant growth and development, lignin is the major plant cell wall component responsible for biomass recalcitrance to industrial processing. The fact that lignin is a non-linear aromatic polymer built with chemically diverse and poorly reactive linkages and a variety of monomer units precludes the ability of any single enzyme to properly recognize and degrade it. Consequently, the use of lignocellulosic feedstock as a renewable and sustainable resource for the production of biofuels and bio-based materials will depend on the identification and characterization of the factors that determine plant biomass recalcitrance, especially the highly complex phenolic polymer lignin. Here, we summarize the current knowledge regarding lignin metabolism in plants, its effect on biomass recalcitrance and the emergent strategies to modify biomass recalcitrance through metabolic engineering of the lignin pathway. In addition, the potential use of sugarcane as a second-generation biofuel crop and the advances in lignin-related studies in sugarcane are discussed.
biopolymer; genetic engineering; phenylpropanoids; wall components
REVIEW PAPER
An overview of lignin metabolism and its effect on biomass recalcitrance
Igor Cesarino; Pedro Araújo; Adilson Pereira Domingues Júnior; Paulo Mazzafera* * Corresponding author: pmazza@unicamp.br.
Universidade Estadual de Campinas, Instituto de Biologia, Departamento de Biologia Vegetal, Caixa Postal 6109, 13083-970 Campinas, SP, Brazil
ABSTRACT
Lignin, after cellulose, is the second most abundant biopolymer on Earth, accounting for 30% of the organic carbon in the biosphere. It is considered an important evolutionary adaptation of plants during their transition from the aquatic environment to land, since it bestowed the early tracheophytes with physical support to stand upright and enabled long-distance transport of water and solutes by waterproofing the vascular tissue. Although essential for plant growth and development, lignin is the major plant cell wall component responsible for biomass recalcitrance to industrial processing. The fact that lignin is a non-linear aromatic polymer built with chemically diverse and poorly reactive linkages and a variety of monomer units precludes the ability of any single enzyme to properly recognize and degrade it. Consequently, the use of lignocellulosic feedstock as a renewable and sustainable resource for the production of biofuels and bio-based materials will depend on the identification and characterization of the factors that determine plant biomass recalcitrance, especially the highly complex phenolic polymer lignin. Here, we summarize the current knowledge regarding lignin metabolism in plants, its effect on biomass recalcitrance and the emergent strategies to modify biomass recalcitrance through metabolic engineering of the lignin pathway. In addition, the potential use of sugarcane as a second-generation biofuel crop and the advances in lignin-related studies in sugarcane are discussed.
Key words: biopolymer, genetic engineering, phenylpropanoids, wall components
INTRODUCTION
Lignin evolution, biosynthesis and distribution
Around 450 million years ago, during the Late Ordovician, the first plant species started to occupy a challenging new niche, the terrestrial environment (Ferrer et al. 2008). As a consequence, pioneering land plants had to cope with several major stresses, including lack of structural support, harmful UV-B radiation, desiccation stress and potential co-evolving pathogens and herbivores (Weng & Chapple 2010). Among the physiological adaptations that arose during the evolution of early land plants, the emergence of the phenylpropanoid pathway was probably the most crucial evolutionary step. The ability to accumulate simple phenylpropanoids with absorbance maxima in the UV-B range allowed early land plants to resist UV irradiation and, consequently, made survival on land possible (Weng & Chapple 2010). Phenylpropanoids play a role in all aspects of plant responses towards abiotic and biotic stimuli, from mediating plant resistance towards pests to promoting a successful reproduction (Vogt 2010). Although the initial move of early plants onto land was facilitated by the acquisition of the phenylpropanoid metabolism, the true dominance of the terrestrial ecosystem by plants was made possible only with the development of cell walls reinforced with the phenylpropanoid polymer lignin, which bestowed the early tracheophytes with physical support to stand upright and enabled long-distance transport of water and solutes by waterproofing the vascular tissue (Boerjan et al. 2003, Weng & Chapple 2010). In addition, the ability of lignin to resist degradation due to its chemically complex structure made this polymer an effective defensive barrier against herbivores and pathogens (Bonawitz & Chapple 2010). Indeed, since plants do not possess a mechanism to degrade lignin, the deposition of lignin represents a non-recoverable investment of carbon and energy and, therefore, might be tightly regulated to guarantee that carbon skeletons requirement does not outpace availability (Rogers et al. 2005).
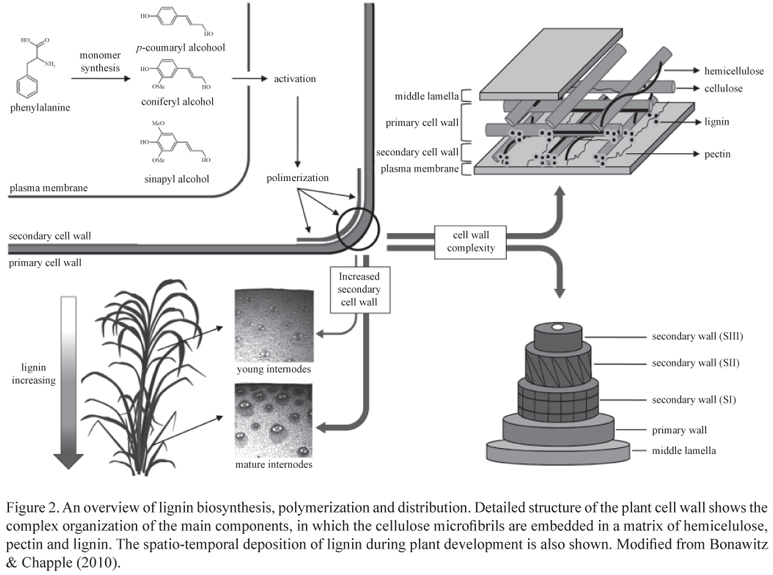
Lignin is a complex racemic heteropolymer produced by the oxidative combinatorial coupling of mainly three p-hydroxycinnamoyl alcohol monomers, the so-called monolignols. The shikimate pathway provides the aromatic amino acid phenylalanine that constitutes the entry point of the monolignol biosynthesis (figure 1). After deamination of phenylalanine by the enzyme phenylalanine ammonia lyase (PAL), aromatic-ring modifications through hydroxylation and methylation followed by the transformation of the carboxylic moiety of the propane tail via esterification and reduction ultimately produce the three p-hydroxycinnamoyl alcohols, p-coumaryl, coniferyl and sinapyl alcohols, that differ in their degree of methoxylation (Liu 2012).
When incorporated into the polymer, these monomers are called p-hydroxyphenyl (H), guaiacyl (G) and syringyl (S) units, respectively, and their individual contribution to lignin composition varies significantly among cell types, taxa and between tissues in the same plant. Phenylpropanoids other than the three canonical monolignols can also be incorporated into the polymer at varying levels, including hydroxycinnamates, hydroxycinnamyl acetates and hydroxycinnamyl aldehydes (Raes et al. 2003). This biopolymer is mainly deposited in the secondarily thickened cell walls of tracheary elements and fibers and, together with hemicellulose, forms a complex matrix in which cellulose microfibrils are embedded (Vega-Sanchez & Ronald 2010, figure 2). The deposition of lignin not only follows a developmental program but can also be triggered by several biotic and abiotic stresses (Moura et al. 2010).
In general, the lignin of angiosperm dicotyledonous plants is composed of G and S units and traces of H units, whereas the lignin from non-flowering vascular plants (i.e. lycophytes, ferns and gymnosperms) is mostly composed of G units with minor amounts of H units. Monocotyledonous lignin contains similar levels of both S and G units and the amount of H-units is relatively higher (Vanholme et al. 2010a). Additionally, grass lignin contains significant amounts of the hydroxycinnamates ferulic acid and p-coumaric acid (Vogel 2008). Non-vascular organisms do not synthesize lignin, although bryophytes can accumulate soluble phenylpropanoids like flavonoids and lignans (Weng & Chapple 2010). Despite this proposed evolutionary scenario, notable exceptions were found in recent years including the discovery of lignin in peripheral cells of the red algae Calliarthron cheilosporioides M. (Martone et al. 2009), the presence of lignin in non-vascular tissues of the liverwort Marchantia polymorpha L. (Espineira et al. 2011) and the recruitment of sinapyl alcohol for S lignin biosynthesis in suspension cell cultures from the basal gymnosperm Ginkgo biloba L. (Uzal et al. 2009).
Cell wall lignification, as well as the incorporation of each monomer, is regulated in a spatio-temporal fashion and varies between primary and secondary cell walls and among tissues (Grabber et al. 2004). Lignin content increases with plant maturity. The incorporation of H and G units starts at the onset of lignification, while only a few S units are detected during early stages of lignin deposition. Subsequently, coniferyl alcohol and increasing amounts of sinapyl alcohol are incorporated to form a mix of G and S units during secondary wall formation (Grabber 2005). In grass cell walls, hydroxycinnamates are also incorporated during secondary cell wall development and lignification. While ferulic acid is the major hydroxycinnamic derivative in young grass cell walls, p-coumaric acid is an indicator of cell wall maturity, since it is mainly esterified to side chains of S units and its incorporation follows the same deposition pattern of syringyl units (Riboulet et al. 2009). Differential distribution of lignin monomers is also observed in case of specific cell types. In most dicotyledonous plants, thickening of secondary walls occurs mainly in water-conducing cells of the xylem, which tend to contain higher levels of G units, and in structural fibers, which are typically enriched in S units (Bonawitz & Chapple 2010, figure 2). In grasses, parenchyma and other tissues such as epidermis and hypodermis present limited but significant lignification and xylary tissues and sclerenchyma accumulate high amounts of lignin and are highly enriched in S lignins (Grabber et al. 2004). The ratio of S/G subunits in lignin also predicts the degree and nature of polymeric cross-linking. G-rich tissues have a more highly cross-linked lignin featuring a greater proportion of biphenyl and other carbon-carbon bonds, whereas S-rich lignin is less condensed, linked through more labile ether bonds at the 4-hydroxyl position (Ferrer et al. 2008). Additional methoxy groups on a lignin monomer results in less possible combinations during polymerization due to reduced available reactive sites. Consequently, S-rich lignin is more easily depolymerized than lignin rich in G units (Ziebell et al. 2010).
Monolignol transport and polymerization
Monolignols are synthesized in the cytoplasm and transported to the cell wall, where they are oxidized prior to their incorporation into the polymer (Vanholme et al. 2008). In contrast to the extensive knowledge of monolignol biosynthesis, little is known about the precise molecular mechanism of monolignol transport towards the cell wall. At least three systems were proposed: i) exocytosis through ER-Golgi network; ii) passive diffusion through plasma membrane; and iii) active transport via specific membrane-localized transporters (Liu 2012). Nevertheless, recent biochemical and genetic data strongly support the third hypothesis. By using isolated plasma and vacuolar membrane vesicles from Arabidopsis thaliana and different transporter inhibitors, Miao & Liu (2010) revealed several lines of evidence of the involvement of ABC-like transporters in monolignols transport across the plasmalemma and their sequestration into the vacuoles: 1) both processes were largely dependent on the presence ATP, which suggest that they are active processes; 2) uptake activity of both vacuolar and plasma membrane vesicles was strongly reduced by specific ABC transporter inhibitors in the presence of ATP; 3) the uptake process displays a typical protein-ligand binding kinetics, excluding the possibility of a passive diffusion process; and 4) vacuolar transporters presented high selectivity for monolignol glucoconjugates, whereas plasma membrane transporters preferentially transport monolignol aglycones (Miao & Liu 2010, Liu 2012). In another study, a role for an ATP-binding cassette transporter in p-coumaryl alcohol transport in Arabidopsis thaliana was demonstrated (Alejandro et al. 2012). Yeast cells expressing AtABCG29 exhibited increased tolerance to p-coumaryl alcohol while Arabidopsis loss-of-function mutants contained less lignin subunits and were more sensitive to p-coumaryl alcohol. This was the first identification of a monolignol transporter, although the characterization of specific transporters for coniferyl and sinapyl alcohol awaits further investigation.
After exported to the cell wall, monolignols are oxidized by peroxidases and/or laccases and further undergo polymerization through combinatorial radical-radical coupling (Morreel et al. 2010a). Both types of enzymes are present as large multigene families with redundant members, which make the study of lignin polymerization process difficult since generation of transgenic plants often results in no visible phenotype. In addition, their broad in vitro substrate specificity cannot offer precise information about their real substrates in planta (McCaig et al. 2005, Cosio & Dunand 2009). Nevertheless, the involvement of peroxidases in lignin polymerization has been clearly demonstrated by a small number of studies reporting the generation of transgenic plants in which lignification was affected by the up- or down-regulation of a particular peroxidase gene (Marjamaa et al. 2009, Fagerstedt et al. 2010). By contrast, only recently the role of laccases in lignification of stems was unambiguously demonstrated by reverse genetics. Berthet et al. (2011) demonstrated that disruption of AtLAC4 and AtLAC17 resulted in tissue-specific alterations to lignification in Arabidopsis thaliana stems. Still, genetic evidence for the functions of individual peroxidase or laccase genes in plants is mostly lacking, especially for monocotyledonous plants.
Following the oxidation, the resulting electron-delocalized monolignol radical presents unpaired electron density at the positions 1-, 3-, O-4-, 5- and 8- and, as the radical coupling is favored at the 8-position, coupling with another monolignol radical results in a mixture of dehydrodimers 8-5-, 8-8- and 8-O-4-linkages (Morreel et al. 2010b). After dimerization, the polymerization process will continue by the coupling of O-4-position of the dimer's phenolic end to the 8-position of the incoming monolignol radical (Morreel et al. 2010a). Lignin is further produced by the addition of monolignols to the growing polymer and not by the concatenation of preformed oligolignols (Morreel et al. 2004). Because lignification is a strictly chemical process, any phenol localized in the lignification site of the cell wall can be used in the combinatorial radical coupling process, influenced by simple physical parameters such as temperature, pH, monolignol supply, concentration of hydrogen peroxide and oxidative enzymes and interactions with polysaccharides present in the matrix (Vanholme et al. 2008). Recently, a new hypothesis suggesting a strong biological control over the phenoxy radical coupling by the so-called dirigent proteins was proposed (Davin & Lewis 2005). However, current knowledge based on genetic data fall clearly in line with the chemical combinatory model (Bonawitz & Chapple 2010).
Lignin and the recalcitrance of plant biomass
Currently, the major bioenergy products are bioethanol, biodiesel and biogas. The bioethanol is produced through the fermentation of soluble sugars or starch, predominantly from sugarcane and maize kernel, and is often referred to as first-generation bioethanol (Yuan et al. 2008). An emerging effort has been expended to increase the use of plant lignocellulosic biomass for the production of bioethanol, the so-called second generation bioethanol (Carroll & Somerville 2009). Lignocellulosic biomass is composed of the polymers cellulose, hemicelluloses and lignin and is a renewable resource for the production of biofuels and bio-based materials (Vanholme et al. 2010c). However, lignocellulose presents a highly complex nature due to its molecular structure and heterogeneity, in which cellulose microfibrils are embedded in a matrix of hemicellulosic polysaccharides covalently cross-linked with the heterogenic and complex lignin (Vega-Sanchez & Ronald 2010). The recalcitrant nature of this mixture, mainly caused by the ability of lignin to resist enzymatic degradation, is one of the major obstacles to convert plant cell wall polysaccharides into fermentable sugars for biofuels production (Vanholme et al. 2010c). The fact that lignin is a non-linear polymer built with chemically diverse and poorly reactive linkages and a variety of monomer units, makes this phenolic polymer the major plant cell wall component responsible for biomass recalcitrance (Weng et al. 2008). Consequently, the conversion of biomass to biofuel requires costly and harsh pre-treatments to degrade lignin and further allow the access to polysaccharides for saccharification (Simmons et al. 2010).
Metabolic engineering of lignin to modify biomass recalcitrance
An emergent strategy in the biofuel field is the genetic engineering of plants that either accumulate less lignin or produce lignins that are more amenable to chemical degradation, in an attempt to facilitate the processing of plant biomass (Carroll & Somerville 2009). A common approach is to use genetic engineering to modify the expression of the enzymes responsible for lignin biosynthesis (Simmons et al. 2010). The independent downregulation of genes encoding seven enzymes of the monolignol pathway in alfalfa resulted in a reduction of the total flux into lignin, with more drastic effects found in the case of earlier enzymes in the pathway (Chen et al. 2006). In addition, striking differences in the lignin composition was observed dependent upon the targeted gene. While the downregulation of C3H or HCT significantly increased the proportion of H units, the reduction in F5H or COMT expression resulted in lower levels of S units, consistent with the position of these enzymes in the pathway. A subsequent study in which the same transgenic lines were analyzed showed that biomass recalcitrance to both acid pretreatment and enzymatic digestion is directly proportional to lignin content (Chen & Dixon 2007). Noteworthy, the saccharification efficiency of untreated biomass of the HCT and C3H lines, which had the lowest lignin content at < 50% of the wild type, was greater than that of pretreated wild type biomass. However, reducing lignin content to improve polysaccharide accessibility is not the only strategy, since the modification of its monomeric composition can also modify lignocellulose recalcitrance (Simmons et al. 2010). Down-regulation of CAD in lignifying tissues in alfalfa did not result in any reduction of lignin content but showed an improvement in digestibility and pulping process (Baucher et al. 1999). Poplar plants overexpressing F5H showed high levels of S-lignin and therefore digest and pulp more easily (Huntley et al. 2003).
Since drastic reductions in lignin levels have negative effects on plant growth and development but even large shifts in lignin composition are still tolerable, the incorporation of atypical phenolic monomers into lignin through genetic engineering to produce a more easily degradable lignin is a new trend in the field (Vanholme et al. 2012). A remarkable example of such strategy comes from the concomitant down-regulation of COMT and up-regulation of F5H in Arabidopsis (Vanholme et al. 2010b). Both enzymes are required to redirect the flux from coniferyl alcohol precursors towards sinapyl alcohol; the hydroxylation of coniferaldehyde by F5H produces 5-hydroxyconiferaldehyde that in turn is methylated by COMT (Boerjan et al. 2003). Since the hydroxylation step performed by F5H is essentially irreversible and 5-hydroxyconiferaldehyde cannot be converted to sinapaldehyde due to COMT down-regulation, CAD catalyzes the convertion of 5-hydroxyconiferaldehyde to 5-hydroxyconiferalcohol. This novel monomer behaves identically as a canonical monolignol, being exported to the cell wall and cross-coupling with normal lignin units to produce a polymer composed almost entirely from units that are undetectable in wild type plants (Vanholme et al. 2010b). Therefore, the incorporation of "monolignol-substitutes" that can introduce readily cleavable linkages into the complex structure of lignin polymer seems feasible and can be a convenient option to achieve a more effective deconstruction of the plant cell wall (Simmons et al. 2010, Vanholme et al. 2012).
Sugarcane as bioenergy crop
Sugarcane ethanol program in Brazil is the most successful example of efficient use of renewable energy (Goldemberg 2007). Brazil is the world's largest sugarcane producer and the leading country in ethanol production. In the 2009/2010 crop season approximately 612 million tons of sugarcane were produced, from which 25 billion liters of ethanol were generated (Cheavegatti-Gianotto et al. 2011). Sugarcane is a prominent bioenergy crop due to the ability to accumulate up to 50-60% of dry weight of stem as sucrose in the mature tissues, a unique feature within Poaceae (Casu et al. 2004). Although the production of sugar-based ethanol represents the most convenient option for sustainable energy in the case of sugarcane, lignocellulosic biomass can also be used, especially because sugarcane is among the most efficient biomass producers known (Vermerris 2011). Sugarcane bagasse, the residue produced after sucrose extraction, is a plentiful lignocellulosic feedstock, composed by 39% cellulose, 25% hemicelluloses and 23% lignin, among other minor components (Carroll & Somerville 2009, Rabelo et al. 2011). At present, the recovered bagasse is used for generation of heat and power for sugar processing into ethanol and also in electricity production that is sold into the grid (Carroll & Somerville 2009). Therefore, the sugarcane industry could benefit from the production of second generation ethanol since the bagasse is promptly available after sucrose extraction and cellulosic ethanol could be co-produced and share part of the infrastructure used for the production of first generation ethanol (Dias et al. 2009, 2012, Rabelo et al. 2011). Most of our current knowledge of lignin metabolism is derived from studies in dicotyledonous herbaceous plants like Arabidopsis and alfalfa, but the mechanisms underlying lignin biosynthesis, polymerization and regulation are not necessarily conserved among all vascular plants (Li et al. 2008). In addition, bioenergy crops may ultimately include dicotyledonous woody plants, such as eucalyptus and hybrid poplar, and monocotyledonous grasses, such as sugarcane and switchgrass (Weng et al. 2008). For instance, genetic information on lignin biosynthesis in sugarcane is scarce and limited to few studies reporting general expression profiling in which some phenylpropanoid genes were differentially expressed during sugarcane stem development or in relation to sucrose accumulation (Casu et al. 2007, Papini-Terzi et al. 2009). Only recently, the first attempts to perform functional characterization of lignin-related genes in sugarcane were reported. The down-regulation of a sugarcane COMT gene reduced lignin content by up to 14%, resulting in an increase of 29% in saccharification yield (Jung et al. 2012). In addition, evidence for the involvement of specific peroxidase isoforms in the lignification process were obtained by analyzing the activity, proteomic profile and expression pattern of class III peroxidases in sugarcane stems (Cesarino et al. 2012b) and suspension cell cultures (Cesarino et al. 2012a). However, despite these recent advances, there is still much to be explored and determined in terms of gene expression and pathway regulation of lignin biosynthesis in sugarcane. In addition, genetic transformation of sugarcane has continued to be a major bottleneck, hampering the use of reverse genetics to study gene function (Arruda 2012). Thus, a deeper understanding of biomass recalcitrance, and therefore of lignin biosynthesis and regulation, is required in order to develop sugarcane varieties that are more amenable to biomass processing and, consequently, to fully benefit from the potential of cellulosic biofuels.
Acknowledgments This work was financially supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (Fapesp, grant BIOEN 08/58035-6). The authors thank Capes-Brazil (I.C. and P.A.) and Fapesp (A.P.D.J.) for doctoral fellowships and CNPq-Brazil for research fellowship (P.M.).
(received: November 07, 2012; accepted: November 14, 2012)
- Alejandro S, Lee Y, Tohge T, Sudre D, Osorio S, Park J, Bovet L, Geldner N, Fernie AR, Martinoia E. 2012. AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Current Biology 22:1207-1212.
- Arruda P. 2012. Genetically modified sugarcane for bioenergy generation. Current Opinion in Biotechnology 23:315-322.
- Baucher M, Bernard-vailhé MA, Chabbert B, Besle J-M, Opsomer C, Van Montagu M, Botterman J. 1999. Down-regulation of cinnamyl alcohol dehydrogenase in transgenic alfalfa (Medicago sativa L.) and the effect on lignin composition and digestibility. Plant Molecular Biology 39:437-447.
- Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cezard L, Le Bris P, Borrega N, Herve J, Blondet E, Balzergue S, Lapierre C, Jouanin L. 2011. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23:1124-1137.
- Boerjan W, Ralph J, Baucher M. 2003. Lignin biosynthesis. Annual Review of Plant Biology 54:519-546.
- Bonawitz ND, Chapple C. 2010. The genetics of lignin biosynthesis: connecting genotype to phenotype. Annual Review of Genetics 44:337-363.
- Carroll A, Somerville C. 2009. Cellulosic biofuels. Annual Review of Plant Biology 60:165-182.
- Casu RE, Dimmock CM, Chapman SC, Grof CP, McIntyre CL, Bonnett GD, Manners JM. 2004. Identification of differentially expressed transcripts from maturing stem of sugarcane by in silico analysis of stem expressed sequence tags and gene expression profiling. Plant Molecular Biology 54:503-517.
- Casu RE, Jarmey JM, Bonnett GD, Manners JM. 2007. Identification of transcripts associated with cell wall metabolism and development in the stem of sugarcane by Affymetrix GeneChip Sugarcane Genome Array expression profiling. Functional and Integrative Genomics 7:153-167.
- Cesarino I, Araújo P, Paes Leme AF, Creste S, Mazzafera P. 2012a. Suspension cell culture as a tool for the characterization of class III peroxidases in sugarcane. Plant Physiology and Biochemistry DOI: 10.1016/j.plaphy.2012.10.015.
- Cesarino I, Araújo P, Sampaio Mayer JL, Paes Leme AF, Mazzafera P. 2012b. Enzymatic activity and proteomic profile of class III peroxidases during sugarcane stem development. Plant Physiology and Biochemistry 55:66-76.
- Cheavegatti-Gianotto A, de Abreu HMC, Arruda P, Bespalhok Filho JC, Burnquist WL, Creste S, di Ciero L, Ferro JA, de Oliveira Figueira AV, Filgueiras TS, Grossi-de-Sá MF, Guzzo EC, Hoffmann HP, de Andrade Landell MG, Macedo N, Matsuoka S, de Castro Reinach F, Romano E, da Silva WJ, de Castro Silva Filho M, Cesar Ulian E. 2011. Sugarcane (Saccharum × officinarum): a reference study for the regulation of genetically modified cultivars in Brazil. Tropical Plant Biology 4:62-89.
- Chen F, Dixon RA. 2007. Lignin modification improves fermentable sugar yields for biofuel production. Nature Biotechnology 25:759-761.
- Chen F, Srinivasa Reddy MS, Temple S, Jackson L, Shadle G, Dixon RA. 2006. Multi-site genetic modulation of monolignol biosynthesis suggests new routes for formation of syringyl lignin and wall-bound ferulic acid in alfalfa (Medicago sativa L.). Plant Journal 48:113-124.
- Cosio C, Dunand C. 2009. Specific functions of individual class III peroxidase genes. Journal of Experimental Botany 60:391-408.
- Davin LB, Lewis NG. 2005. Lignin primary structures and dirigent sites. Current Opinion in Biotechnology 16:407-415.
- Dias MO, Junqueira TL, Cavalett O, Cunha MP, Jesus CD, Rossell CE, Maciel Filho R, Bonomi A. 2012. Integrated versus stand-alone second generation ethanol production from sugarcane bagasse and trash. Bioresource Technology 103:152-161.
- Dias MOS, Ensinas AV, Nebra SA, Maciel R, Rossell CEV, Maciel MRW. 2009. Production of bioethanol and other bio-based materials from sugarcane bagasse: Integration to conventional bioethanol production process. Chemical Engineering Research & Design 87:1206-1216.
- Espineira JM, Novo Uzal E, Gomez Ros LV, Carrion JS, Merino F, Ros Barcelo A, Pomar F. 2011. Distribution of lignin monomers and the evolution of lignification among lower plants. Plant Biology 13:59-68.
- Fagerstedt KV, Kukkola EM, Koistinen VV, Takahashi J, Marjamaa K. 2010. Cell wall lignin is polymerised by class III secretable plant peroxidases in Norway spruce. Journal of Integrative Plant Biology 52:186-194.
- Ferrer JL, Austin MB, Stewart Junior C, Noel JP. 2008. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiology and Biochemistry 46:356-370.
- Goldemberg J. 2007. Ethanol for a sustainable energy future. Science 315:808-810.
- Grabber JH. 2005. How do lignin composition, structure, and cross-linking affect degradability? A review of cell wall model studies. Crop Science 45:820-831.
- Grabber JH, Ralph J, Lapierre C, Barriere Y. 2004. Genetic and molecular basis of grass cell-wall degradability. I. Lignin-cell wall matrix interactions. Comptes Rendus Biologies 327:455-465.
- Huntley SK, Ellis D, Gilbert M, Chapple C, Mansfield SD. 2003. Significant increases in pulping efficiency in C4H-F5H-transformed poplars: improved chemical savings and reduced environmental toxins. Journal of Agricultural and Food Chemistry 51:6178-6183.
- Jung JH, Fouad WM, Vermerris W, Gallo M, Altpeter F. 2012. RNAi suppression of lignin biosynthesis in sugarcane reduces recalcitrance for biofuel production from lignocellulosic biomass. Plant Biotechnology Journal 10:1067-1076.
- Li X, Weng JK, Chapple C. 2008. Improvement of biomass through lignin modification. Plant Journal 54:569-581.
- Liu CJ. 2012. Deciphering the enigma of lignification: precursor transport, oxidation, and the topochemistry of lignin assembly. Molecular Plant 5:304-317.
- Marjamaa K, Kukkola EM, Fagerstedt KV. 2009. The role of xylem class III peroxidases in lignification. Journal of Experimental Botany 60:367-376.
- Martone PT, Estevez JM, Lu F, Ruel K, Denny MW, Somerville C, Ralph J. 2009. Discovery of lignin in seaweed reveals convergent evolution of cell-wall architecture. Current Biology 19:169-175.
- McCaig BC, Meagher RB, Dean JF. 2005. Gene structure and molecular analysis of the laccase-like multicopper oxidase (LMCO) gene family in Arabidopsis thaliana Planta 221:619-636.
- Miao YC, Liu CJ. 2010. ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proccedings of the National Academy of Science of the United States of America 107:22728-22733.
- Morreel K, Dima O, Kim H, Lu F, Niculaes C, Vanholme R, Dauwe R, Goeminne G, Inze D, Messens E, Ralph J, Boerjan W. 2010a. Mass spectrometry-based sequencing of lignin oligomers. Plant Physiology 153:1464-1478.
- Morreel K, Kim H, Lu F, Dima O, Akiyama T, Vanholme R, Niculaes C, Goeminne G, Inze D, Messens E, Ralph J, Boerjan W, 2010b. Mass spectrometry-based fragmentation as an identification tool in lignomics. Analytical Chemistry 82:8095-8105.
- Morreel K, Ralph J, Kim H, Lu F, Goeminne G, Ralph S, Messens E, Boerjan W. 2004. Profiling of oligolignols reveals monolignol coupling conditions in lignifying poplar xylem. Plant Physiology 136:3537-3549.
- Moura JCMS, Bonine CAV, Viana JDF, Dornelas MC, Mazzafera P. 2010. Abiotic and biotic stresses and changes in the lignin content and composition in plants. Journal of Integrative Plant Biology 52:360-376.
- Papini-Terzi FS, Rocha FR, Vencio RZ, Felix JM, Branco DS, Waclawovsky AJ, Del Bem LE, Lembke CG, Costa MD, Nishiyama Junior MY, Vicentini R, Vincentz MG, Ulian EC, Menossi M, Souza GM. 2009. Sugarcane genes associated with sucrose content. BMC Genomics 10:120.
- Rabelo SC, Carrere H, Maciel Filho R, Costa AC. 2011. Production of bioethanol, methane and heat from sugarcane bagasse in a biorefinery concept. Bioresource Technology 102:7887-7895.
- Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W. 2003. Genome-wide characterization of the lignification toolbox in Arabidopsis Plant Physiology 133:1051-1071.
- Riboulet C, Guillaumie S, Mechin V, Bosio M, Pichon M, Goffner D, Lapierre C, Pollet B, Lefevre B, Martinant JP, Barriere Y. 2009. Kinetics of phenylpropanoid gene expression in maize growing internodes: relationships with cell wall deposition. Crop Science 49:211-223.
- Rogers LA, Dubos C, Cullis IF, Surman C, Poole M, Willment J, Mansfield SD, Campbell MM. 2005. Light, the circadian clock, and sugar perception in the control of lignin biosynthesis. Journal of Experimental Botany 56:1651-1663.
- Simmons BA, Loque D, Ralph J. 2010. Advances in modifying lignin for enhanced biofuel production. Current Opinion in Plant Biology 13:313-320.
- Uzal EN, Gomez Ros LV, Pomar F, Bernal MA, Paradela A, Albar JP, Ros Barcelo A, 2009. The presence of sinapyl lignin in Ginkgo biloba cell cultures changes our views of the evolution of lignin biosynthesis. Physiologia Plantarum 135:196-213.
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W. 2010a. Lignin biosynthesis and structure. Plant Physiology 153:895-905.
- Vanholme R, Morreel K, Darrah C, Oyarce P, Grabber JH, Ralph J, Boerjan W. 2012. Metabolic engineering of novel lignin in biomass crops. New Phytologist 196:978-1000.
- Vanholme R, Morreel K, Ralph J, Boerjan W. 2008. Lignin engineering. Current Opinion in Plant Biology 11:278-285.
- Vanholme R, Ralph J, Akiyama T, Lu F, Pazo JR, Kim H, Christensen JH, Van Reusel B, Storme V, De Rycke R, Rohde A, Morreel K, Boerjan W. 2010b. Engineering traditional monolignols out of lignin by concomitant up-regulation of F5H1 and down-regulation of COMT in Arabidopsis Plant Journal 64:885-897.
- Vanholme R, Van Acker R, Boerjan W. 2010c. Potential of Arabidopsis systems biology to advance the biofuel field. Trends Biotechnology 28:543-547.
- Vega-Sanchez ME, Ronald PC. 2010. Genetic and biotechnological approaches for biofuel crop improvement. Current Opinion in Biotechnology 21:218-224.
- Vermerris W. 2011. Survey of genomics approaches to improve bioenergy traits in maize, sorghum and sugarcane. Journal of Integrative Plant Biology 53:105-119.
- Vogel J. 2008. Unique aspects of the grass cell wall. Current Opinion of Plant Biology 11:301-307.
- Vogt T. 2010. Phenylpropanoid biosynthesis. Molecular Plant 3:2-20.
- Weng JK, Chapple C. 2010. The origin and evolution of lignin biosynthesis. New Phytologist 187:273-285.
- Weng JK, Li X, Bonawitz ND, Chapple C. 2008. Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Current Opinion in Biotechnology 19:166-172.
- Yuan JS, Tiller KH, Al-Ahmad H, Stewart NR, Stewart Junior CN. 2008. Plants to power: bioenergy to fuel the future. Trends in Plant Science 13:421-429.
- Ziebell A, Gracom K, Katahira R, Chen F, Pu Y, Ragauskas A, Dixon RA, Davis M. 2010. Increase in 4-coumaryl alcohol units during lignification in alfalfa (Medicago sativa) alters the extractability and molecular weight of lignin. Journal of Biological Chemistry 285:38961-38968.
Publication Dates
-
Publication in this collection
04 Jan 2013 -
Date of issue
2012
History
-
Received
07 Nov 2012 -
Accepted
14 Nov 2012