Abstract
The inducible tetracycline resistance determinant isolated from Proteus mirabilis cloned into the plasmid pACYC177 was mutagenized by insertion of a mini-Mu-lac phage in order to define the regions in the cloned sequences encoding the structural and regulatory proteins. Three different types of mutants were obtained: one lost the resistance phenotype and became Lac+; another expressed the resistance at lower levels and constitutively; the third was still dependent on induction but showed a lower minimal inhibitory concentration. The mutant phenotypes and the locations of the insertions indicate that the determinant is composed of a repressor gene and a structural gene which are not transcribed divergently as are other known tetracycline determinants isolated from Gram-negative bacteria
tetracycline; mutation; lacZ-fusion; Proteus mirabilis; bacteriophage Mu
Braz J Med Biol Res, March 1997, Volume 30(3) 363-367 (Short Communication)
Mini-Mu insertions in the tetracycline resistance determinant from Proteus mirabilis
V.D. Magalhães and B.A. Castilho
Departamento de Microbiologia, Imunologia e Parasitologia, Escola Paulista de Medicina, Universidade Federal de São Paulo, 04023-062 São Paulo, SP, Brasil
 Text
Text
 References
References
 Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Abstract
The inducible tetracycline resistance determinant isolated from Proteus mirabilis cloned into the plasmid pACYC177 was mutagenized by insertion of a mini-Mu-lac phage in order to define the regions in the cloned sequences encoding the structural and regulatory proteins. Three different types of mutants were obtained: one lost the resistance phenotype and became Lac+; another expressed the resistance at lower levels and constitutively; the third was still dependent on induction but showed a lower minimal inhibitory concentration. The mutant phenotypes and the locations of the insertions indicate that the determinant is composed of a repressor gene and a structural gene which are not transcribed divergently as are other known tetracycline determinants isolated from Gram-negative bacteria.
Key words: tetracycline, mutation, lacZ-fusion, Proteus mirabilis, bacteriophage Mu
The tetracycline resistance (TcR) determinants isolated from Gram-negative bacteria have been classified from A through H based on hybridization experiments and nucleotide sequencing data (1,2). The majority of TcR determinants share the same genetic organization - a gene encoding the tetracycline resistance structural protein (tetA) and another encoding the repressor (tetR). The two genes are transcribed divergently from a region where consensus sequences of overlapping promoters and two palindromic operator sequences were described (3). The resistance phenotype is inducible by subinhibitory concentrations of tetracycline (4,5).
The resistance protein is localized in the cytoplasmic membrane and promotes the efflux of the drug. It has been described as a highly hydrophobic protein with 12 transmembrane domains in a-helical conformation (6). The primary structure of the protein, deduced from nucleotide sequencing, revealed a prominent hydrophilic region located in the center of the polypeptide chain, which is also a feature of other integral membrane proteins with transport function (6). Point and deletion mutations of the resistance protein characterized two complementation groups, which define two domains called teta and tetß (7). There is evidence that the efflux of tetracycline is driven by a multimeric form of the TetA protein (8).
The repressor protein negatively regulates its own synthesis as well as that of the resistance membrane protein (5). Tetracycline, complexed with a divalent metal ion (Mg2+), functions as an inducer of the system (3). The drug has a much stronger affinity for TetR than for the ribosome, and the repressor also binds preferentially to tetracycline rather than to the operator region (9). The binding of tetracycline to the two repressor dimers, that cooperatively occupy both operators, induces conformational changes in TetR, hindering the interaction with DNA (10). Mutants of the repressor show a lower level of resistance, and a constitutive phenotype. The former observation has led to the hypothesis that TetR may participate in the mechanism of resistance mediated by TetA (11,12).
P. mirabilis expresses an inducible chromosomal tetracycline resistance that does not hybridize to the already described systems (1). This determinant has been isolated as part of a 2.1-kb DNA fragment obtained by partial Sau3A digestion of P. mirabilis chromosomal DNA and cloned at the BamHI site of the medium copy number plasmid pACYC177 (ApR, KmR) (13), where ApR indicates ampicillin resistance and KmR indicates kanamycin resistance, originating plasmid pVM3. This fragment confers on the E. coli host a fully inducible resistance to tetracycline. The cloned determinant has also been characterized as promoting the efflux of the drug (Magalhães VD, Schumann W and Castilho BA, unpublished data).
We describe here the use of a transposable element, the mini-Mu phage MudII1734 (14), to mutagenize the fragment carrying the TcR determinant sequences of P. mirabilis in order to locate both the repressor and the structural resistance protein genes. This method was chosen because the cloned sequence possesses few restriction sites suitable for subcloning to aid in the precise localization of these genes. This information was important both for sequencing a limited region of DNA and later for assigning an open reading frame to its function.
The MudII1734 phage lacks the genes involved in transposition/replication and lytic functions; these can be provided in trans by a helper phage (Mucts), which codes for a temperature-sensitive repressor. In addition, MudII1734 carries a kanamycin resistance marker (KmR) for selection and contains the lac operon structural genes, without the promoter or the translation initiation signal of the lacZ gene. Transcription and translation initiated in bacterial sequences outside the right end of the phage can proceed across the Mu sequences, and, provided that the correct reading frame is used, result in the expression of ß-galactosidase activity. Thus, mutagenesis with this phage allows the localization of promoters and of translated sequences. MudII1734 has 9.5 kb and a unique HindIII recognition site located 1.0 kb from the left end. It also has two recognition sites for BamHI, 0.1 kb from the right end and 2.7 kb from the left end.
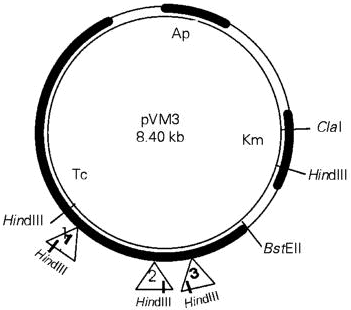
Because the pVM3 plasmid confers resistance on kanamycin, derived from pACYC177, and this was also the marker carried by MudII1734, the sequences of the vector responsible for this phenotype had to be eliminated. A 7.2-kb ClaI-BstEII fragment of pVM3 was isolated (Figure 1), and its termini were made blunt by treatment with Bal31. It was then circularized by ligation in the presence of a BamHI linker. The resulting plasmid, called pVM383, contains the complete P. mirabilis-derived fragment conferring resistance to tetracycline.
- Structure of the plasmid pVM3 and the location of mini-Mu insertions. A partial restriction map of the original plasmid pVM3 is shown with the recognition sites for relevant enzymes. The locations of the MudII1734 insertions (triangles) in plasmid pVM383, derived from pVM3 after removing a 7.2-kb ClaI-BstEII fragment, deduced from BamHI and HindIII digestion of the mutated plasmids, are also shown, since the P. mirabilis sequences were maintained in both plasmids. 1, pVM27; 2, pVM77; 3, pVM91. The HindIII recognition site at the left end of MudII1734 is shown to indicate the orientation of the insertions. Kanamycin (Km) and ampicillin (Ap) were used, when appropriate, at the concentrations of 50 µg/ml and 100 µg/ml, respectively. Transformation of E. coli cells by the CaCl2 method, plasmid isolation, DNA manipulation methods and gel electrophoresis were performed according to Sambrook et al. (15), where the media are also indicated. Digestion with restriction enzymes was performed as recommended by the manufacturer (GIBCO-BRL). Tc, Tetracycline.
Plasmid pVM383 was then used to transform E. coli POII1734, which harbors the mini-Mu phage MudII1734 and a lysogenic Mucts, with selection for KmR and ApR. In order to isolate random insertions of the mini-Mu specifically into plasmid sequences, a lysate prepared as previously described (14) from POII1734 carrying pVM383 was used to infect Xph43Mucts cells (16), and KmRApR transductants were selected. In this process, after induction, the mini-Mu transposes to several locations, including plasmid sequences; in this case, co-integrated structures can be formed with two copies of the MudII1734 flanking the plasmid molecule. These are packaged in viral particles and upon infection of a recipient strain, homologous recombination can occur, reforming the plasmid molecule now carrying an insertion of the mini-Mu phage. These events can be obtained by applying selection for both the plasmid (ApR) and the mini-Mu (KmR).
The phenotype of the transductants obtained was tested by tooth-picking colonies to Mueller-Hinton (MH) plates containing tetracycline (1 µg/ml, for full induction of resistance) or without the antibiotic (non-induced cells). They were then transferred to McConkey plates supplemented with tetracycline (1 µg/ml) to test the Lac phenotype and to MH plates containing different concentrations of the drug (20, 50 and 100 µg/ml).
Three transductants with different phenotypes (Table 1) had their plasmids named pVM27, pVM77 and pVM91, analyzed by digestion with HindIII and BamHI, followed by agarose gel electrophoresis in order to locate the site of insertion of MudII1734 (Figure 1). This analysis allowed the mapping of the insertions as shown in Figure 1.
Cells carrying plasmid pVM27 show a constitutive low level of tetracycline resistance. This phenotype resembles mutants already described carrying deletions in the repressor gene of other systems, which, together with the constitutive phenotype, have also lower levels of resistance (11,12). Restriction enzyme analysis of this plasmid showed only one band of approximately 8.8 kb after HindIII digestion; this band should be a doublet, not only because there are two recognition sites for this enzyme (one in the P. mirabilis sequence and the other in the mini-Mu), but also because the size of the plasmid plus the mini-Mu is 17 kb. This result suggests an insertion of the phage very close to the HindIII site of the sequence from P. mirabilis. The band pattern generated by BamHI digestion allowed the assignment of the location and orientation of this insertion as shown in Figure 1. Based on the phenotype observed, we may conclude that the mini-Mu was inserted in the repressor gene and that this gene is localized near the HindIII site of the insert in pVM3.
The restriction pattern of plasmid pVM91 indicated that the MudII1734 phage was localized to 1.4 kb from the HindIII site of the cloned fragment, as shown in Figure 1. The cells harboring this plasmid are Lac+, indicating that transcription and translation of lacZ could be driven by sequences outside the Mu phage. Thus, the direction of transcription in this region is probably towards the HindIII site. Loss of the resistance phenotype suggests that the insertion occurred in the tetracycline resistance determinant. Assuming 1.2 kb as the approximate size necessary for coding the structural protein and 0.8 kb for the repressor and regulatory region, as deduced from other TcR determinants (9), the insertion into pVM91 probably occurred in the region coding for the a domain of the TetA protein, a region that is essential for its function.
The third mutated plasmid, pVM77, gave rise to restriction enzyme fragments that located its insertion approximately 1.0 kb from the HindIII site, as shown in Figure 1. This plasmid enabled the host cells to grow in the presence of 50 µg/ml tetracycline, and not at 100 µg/ml as does the wild type. However, the resistance phenotype is still inducible. The lower TcR level presented by cells harboring plasmid pVM77 suggests that the insertion occurred at the carboxyl end of the resistance polypeptide, since this phenotype is consistent with previously described deletions of the ß domain of this protein, which also lead to lower MIC's (7,17). Furthermore, lacZ fusions to the carboxyl terminus of the TetA protein have been reported to still maintain some level of resistance (8).
The data presented here indicate that the P. mirabilis tetracycline resistance determinant cloned in plasmid pVM3 is composed of two genes, one coding for the resistance protein and the other coding for the repressor. Given that transcription and translation in the region that encodes for the resistance protein take place towards the HindIII site of the cloned sequence, as deduced from plasmid pVM91, and that the insertion that abolished regulation in plasmid pVM27 occurred closer to this restriction site than that in pVM91, it is reasonable to assume that the repressor and resistance genes are not transcribed divergently from overlapping promoters, as is the case for other TcR determinants (1). The possibility that these sequences have been shuffled during the Sau3A partial digestion and cloning is highly improbable, because another independently isolated fragment of P. mirabilis chromosome encoding the complete TcR determinant was shown to have a restriction map identical to that of pVM3 in the region that was shown here to carry the determinant (Magalhães VD, Schumann W and Castilho BA, unpublished data). Furthermore, for the determinants characterized so far, the overlapping promoters cannot be split into two independent promoters. Sequencing data in progress will provide further insights into this peculiarly organized TcR determinant.
Address for correspondence: V.D. Magalhães, Laboratório de Biologia Molecular, Hospital Israelita Albert Einstein, Av. Albert Einstein, 627, 05651-901 São Paulo, SP, Brasil.
V.D. Magalhães was the recipient of a CNPq predoctoral fellowship. Publication supported by FAPESP. Received August 31, 1995. Accepted January 6, 1997.
- 1. Levy SB (1988). Tetracycline resistance determinants are widespread. ASM News, 54: 418-421.
- 2. Hansen LM, Mamurry LM, Levy SB & Hirsh DC (1993). A new tetracycline resistance determinant, Tet H, from Pasteurella multocida specifying active efflux of tetracycline. Antimicrobial Agents and Chemotherapy, 37: 2699-2705.
- 3. Hillen W & Schollmeier K (1983). Nucleotide sequence of the Tn10 encoded tetracycline resistance gene. Nucleic Acids Research, 11: 525-539.
- 4. Izaki K, Kiuchi K & Arima K (1966). Specificity and mechanism of tetracycline resistance in a multiple drug resistant strain of Escherichia coli Journal of Bacteriology, 91: 628-633.
- 5. Yang H, Zubay G & Levy SB (1976). Synthesis of an R plasmid protein associated with tetracycline resistance is negatively regulated. Proceedings of the National Academy of Sciences, USA, 73: 1509-1512.
- 6. Eckert B & Beck CF (1989). Topology of the transposon Tn10-encoded tetracycline resistance protein within the inner membrane of Escherichia coli Journal of Biological Chemistry, 264: 11663-11670.
- 7. Curiale MS, McMurry LM & Levy SB (1984). Intracistronic complementation of the tetracycline resistance membrane protein of Tn10 Journal of Bacteriology, 157: 211-217.
- 8. Hickman RK & Levy SB (1988). Evidence that TET protein functions as a multimer in the inner membrane of Escherichia coli Journal of Bacteriology, 170: 1715-1720.
- 9. Hillen W, Schollmeier K & Gatz C (1984). Control of expression of the Tn10-encoded tetracycline resistance operon. II. Interaction of RNA polymerase and TET repressor with the tet operon regulatory region. Journal of Molecular Biology, 172: 185-201.
- 10. Hinrichs W, Kisker C, Düvel M, Müller A, Tovar K, Hillen W & Saenger W (1994). Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance. Science, 264: 418-420.
- 11. Levy SB (1984). Resistance to tetracyclines. Antimicrobial Drug Resistance. Academic Press Inc., New York.
- 12. Heuer C, Hickman RK, Curiale MS, Hillen W & Levy SB (1987). Constitutive expression of tetracycline resistance mediated by a Tn10-like element in Haemophilus parainfluenzae results from a mutation in the repressor gene. Journal of Bacteriology, 169: 990-994.
- 13. Chang ACY & Cohen SN (1978). Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. Journal of Bacteriology, 134: 1141-1156.
- 14. Castilho BA, Olfson P & Casadaban MJ (1984). Plasmid insertion mutagenesis and lac gene fusion with mini-Mu bacteriophage transposons. Journal of Bacteriology, 158: 488-495.
- 15. Sambrook J, Fritsch EF & Maniatis T (1989). Molecular Cloning - A Laboratory Manual 2nd edn. CSH Laboratory Press, New York.
- 16. Brickman E & Beckwith J (1975). Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and 80 transducing phages. Journal of Molecular Biology, 96: 307-316.
- 17. Coleman DC & Foster TJ (1981). Analysis of the reduction in expression of tetracycline resistance determined by transposon Tn10 in the multicopy state. Molecular and General Genetics, 182: 171-177.
- 18. NCCLS (1988). The National Committee for Clinical Laboratory Standards. Dilution methods for antimicrobial susceptibility tests for bacteria that grow aerobically. 2nd edn. Villanova, Wayne, PA.
Correspondence and Footnotes
Publication Dates
-
Publication in this collection
19 Mar 1999 -
Date of issue
Mar 1997
History
-
Accepted
06 Jan 1997 -
Received
31 Aug 1995