Abstract
Insulin and glucagon are the hormonal polypeptides secreted by the B and A cells of the endocrine pancreas, respectively. Their major physiologic effects are regulation of carbohydrate metabolism, but they have opposite effects. Insulin and glucagon have various physiologic roles, in addition to the regulation of carbohydrate metabolism. The physiologic effects of insulin and glucagon on the cell are initiated by the binding of each hormone to receptors on the target cells. Morphologic studies may be useful for relating biochemical, physiologic, and pharmacologic information on the receptors to an anatomic background. Receptor radioautography techniques using radioligands to label specific insulin and glucagon receptors have been successfully applied to many tissues and organs. In this review, current knowledge of the histologic distribution of insulin and glucagon receptors is presented with a brief description of receptor radioautography techniques
radioautography; insulin; glucagon; receptor; distribution
Braz J Med Biol Res, February 1998, Volume 31(2) 243-256
Histologic distribution of insulin and glucagon receptors
M. Watanabe, H. Hayasaki, T. Tamayama and M. Shimada
Department of Anatomy, Osaka Medical College, Takatsuki, Osaka, Japan
 Text
Text
Abstract
References
 Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes

Insulin and glucagon are the hormonal polypeptides secreted by the B and A cells of the endocrine pancreas, respectively. Their major physiologic effects are regulation of carbohydrate metabolism, but they have opposite effects. Insulin and glucagon have various physiologic roles, in addition to the regulation of carbohydrate metabolism. The physiologic effects of insulin and glucagon on the cell are initiated by the binding of each hormone to receptors on the target cells. Morphologic studies may be useful for relating biochemical, physiologic, and pharmacologic information on the receptors to an anatomic background. Receptor radioautography techniques using radioligands to label specific insulin and glucagon receptors have been successfully applied to many tissues and organs. In this review, current knowledge of the histologic distribution of insulin and glucagon receptors is presented with a brief description of receptor radioautography techniques.
Key words: radioautography, insulin, glucagon, receptor, distribution
Introduction
Insulin is a hormone secreted by B cells, and glucagon is secreted by A cells of the pancreas. The two hormones play an important role in carbohydrate metabolism. However, the actions of insulin and glucagon in carbohydrate metabolism are opposite. Furthermore, insulin and glucagon have various physiologic roles in addition to the regulation of carbohydrate metabolism. The physiologic effects of insulin and glucagon on the cell are initiated by the binding of each hormone to target cell receptors. To relate biochemical, physiologic, and pharmacologic information on receptors to an anatomic background, morphologic studies might be important, although microdissection techniques allow the determination of receptor levels in tissues as small as 200 to 500 µm in diameter (1,2). Significant advances in the determination of the histologic distribution of receptors for many hormones have been made in the past two decades, primarily by radioautography. In this review, the insulin and glucagon receptors will be described with emphasis on the histological tissue distribution of these receptors from macroscopic to light microscopic levels determined mainly with the radioautographic technique. The fundamental procedure of macro- and microradioautography for peptide hormone receptors will also be described.
Insulin and insulin receptors
Insulin is only secreted by B cells of the islets of Langerhans of the pancreas. Insulin was crystallized by Abel in 1926 and all amino acid sequences were identified by Sanger in 1955. Hodgkin determined the tertiary structure of the protein in 1969. Insulin is a polypeptide with a molecular mass of about 5800 Da, in which the A chain is linked to the B chain by two disulfide bridges. The binding site of the insulin molecule to the receptor has been clarified (3). When the site at which insulin takes part in binding to the receptor is labeled with radioactive iodine, normal binding ability is lost. Therefore, in investigations on insulin binding to the insulin receptors, TyrA14 located away from the receptor-binding site is labeled with radioactive iodine.
Action of insulin
Insulin plays an important physiologic role, especially in the liver, muscle, and adipose cells, in homeostasis of blood glucose concentration (4). For this reason, the liver, muscle, and fat have been regarded as major target tissues for insulin. However, insulin has also been found to promote cellular growth and proliferation in many cell types in culture. The physiologic effects of insulin are divided into three types in terms of the timing of action (5). 1) Immediate effect of insulin: The effect occurs within several seconds after insulin is administered; transportation of glucose is promoted and phosphorylation and dephosphorylation of enzymes occur. 2) Mid-term action of insulin: The effect of insulin is detected within 5-60 min after insulin administration, including the induction and appearance of the gene encoding the protein. The maximum effects of this action are caused within 3-6 h. 3) Long-term action of insulin: The effect can be detected from several hours to several days later, including the stimulation of DNA synthesis, cell division, and cell differentiation.
It should be noted that the action of insulin is different depending on the amount. For instance, the amount of insulin necessary to inhibit gluconeogenesis is greater than that necessary to inhibit glycogenolysis (4). These actions of insulin begin with the binding of insulin to insulin receptors.
Insulin receptors
The action of insulin starts with binding to the receptors located on the membrane of target cells as is the case for all peptide hormones. The kinetic characteristics of insulin receptors in various types of cells have been clarified using radiolabeled insulin (6). The insulin receptor exists on the membrane of all mammalian cells. The brain cell, which has been assumed to have an insulin-independent organization, is also included among these cells (7,8). The number of receptors varies from 40 for erythrocytes to 200~300 x 103 for adipocytes and hepatocytes. The binding characteristics are complex because of negative cooperativity among the insulin-binding sites, in which binding of one insulin molecule to the receptor prevents binding of the second molecule (9).
The insulin receptor consists of two a subunits containing the site for insulin binding and two ß subunits containing the tyrosine kinase domain; these subunits are connected by disulfide bridges to form a 350 kDa ß-a-a-ß tetramer (10). The insulin receptor derives from a single gene on the short arm of chromosome 19 and consists of 22 exons separated by 21 introns (11). However, two isoforms of the insulin receptor are produced by alternative splicing of exon 11 (12).
The distribution of insulin receptors with or without exon 11 varies among tissues (12,13). The isoform with exon 11 is predominant in the liver, but is not common in muscle. The adipocyte has both isoforms at nearly equal levels. These two isoforms of the insulin receptor exhibit different affinities for insulin. The isoform with exon 11 exhibits higher affinity than the isoform without exon 11 (14,15).
The insulin receptor is a hormone-activated protein tyrosine kinase. Protein tyrosine kinases are enzymes that catalyze the transfer of phosphate groups from adenosine triphosphate to the tyrosyl residues of proteins. Binding of insulin to the binding site on the extracellular portion of the a subunit results in activation of the protein kinase site on the cytoplasmic portion of the ß subunit, which adds phosphate groups to tyrosine residues in the target proteins in the cytoplasm. Following receptor kinase activation, an insulin signal is transmitted through postreceptor signaling pathways (16,17).
Glucagon and glucagon receptors
Glucagon was discovered in the 1930s in crude insulin preparations and was termed the hyperglycemic glycogenolytic factor. Glucagon is a single-chain polypeptide with a molecular mass of 3485 Da which consists of 29 amino acids (4). A single gene encoding preproglucagon has been found on chromosome 2 in humans and rats, and certain intestinal and neural cells express the preproglucagon gene (18,19). Glucagon immunoreactivity has also been reported in certain intestinal (20-22) and neural (23) cells, besides pancreatic A cells, but these cells do not appear to secrete significant quantities of true glucagon under normal circumstances (24). In the intestine, glucagon is secreted in the form of glicentin (consisting of 69 amino acids and referred to as enteroglucagon) and oxyntomodulin (37 amino acids), but not in the form of true glucagon (29 amino acids) (25). Therefore, A cells of the islets of Langerhans of the pancreas are the only source of true glucagon under normal circumstances. The binding site of the glucagon molecule to the receptor has been investigated (26,27). Since normal binding ability of glucagon to the receptors will be lost by labeling the binding sites with radioactive iodine, Tyr10, which is not the receptor-binding site, is labeled with radioactive iodine in experiments carried out to investigate glucagon binding.
Action of glucagon
Glucagon has several effects that are opposite to those of insulin (4). Glucagon raises the blood glucose concentration by stimulating hepatic glycogenolysis and gluconeogenesis. In contrast, a fall in glucagon concentration below basal levels results in a decrease in hepatic glucose production. Glucagon has been shown to play a critical role in the disposition of amino acids by increasing their inward transport, degradation, and conversion into glucose. The stimulatory action of glucagon on hepatic glucose production lasts only 30 to 60 min. In addition to its action on carbohydrate metabolism, glucagon is said to be involved in the regulation of lipolysis (28), but within the physiological range, glucagon has little or no effect on the lipolysis of adipose tissue in humans (29,30). Other reported actions of glucagon include the inhibition of gastric acid secretion and gut motility (31), a positive inotropic effect on the heart (32), a spasmolytic function on the intestinal wall (33), involvement in intra-islet hormone regulation in the pancreas (34-36), and regulation of renal function (37,38).
Glucagon receptor
The effects of glucagon are mediated by the binding of the hormone to a specific receptor (24). The human glucagon receptor is located on chromosome 17 (39). The rat glucagon receptor was cloned and found to belong to the GTP family and cyclase-linked receptors having seven putative transmembrane domains (40,41). The N-terminal extracellular portion of the receptor is required for ligand binding and most of the distal C-terminal tail is not necessary for ligand binding; the absence of the C-terminal tail may slightly increase the receptor-binding affinity for glucagon. The C-terminal tail is also not necessary for adenyl cyclase coupling and, therefore, does not play a direct role in G protein activation by the glucagon receptor (42). A human glucagon receptor has also been cloned from human liver tissue, and it was shown that the human glucagon receptor amino acid sequence had 82% identity with the rat receptor (43). The number of receptors is supposed to be 200 x 103 in hepatocytes (40), and it has been proposed that two types of hepatic glucagon receptors may have different signaling pathways (44). The principal hepatic glucagon receptor is a glycoprotein of 63 kDa (45,46) and there is a second receptor of 33 kDa (45). The existence of two functionally distinct forms of glucagon receptors, a high-affinity form with a Kd of 0.1~1.0 nM comprising 1% of the glucagon-binding sites and a low-affinity form with a Kd of 10~100 nM that makes up 99% of the molecules, has been reported (24).
Methods for investigating the histologic distribution of receptors
Although both immunohistochemistry and radioautography can be used to investigate the distribution of insulin and glucagon receptors, we focus here on the radioautographic technique (47). The body is made up of many organs and each individual organ is functionally and morphologically heterogeneous. It is important that experiments of receptor distribution proceed from the macroscopic to the microscopic level. Macroradioautography including whole body radioautography is suitable for studying the tissue distribution of radiolabeled ligands in the whole animal or in large organs. Microradioautography at the light microscopy level is suitable for the visualization of radiolabeled ligand binding at the cellular level.
For radioautography, two types of labeling techniques for receptors can be used in vivo and in vitro. In in vivo receptor radioautography, tissues and organs are removed from experimental animals that have received radiolabeled ligand injections, and sections are prepared. The sites labeled with the ligand are visualized by radioautography with films or nuclear emulsions. During in vitro procedures, sections of tissues and organs from experimental animals that have not been injected with a radiolabeled ligand are incubated with a radiolabeled ligand. The sections are then exposed to films or nuclear emulsions to detect ligand radioactivity. Since many factors, such as ligand metabolism and tissue barriers, might influence in vivo receptor labeling, the interpretation of these results might be more complicated than that of in vitro labeling. However, in vivo procedures might better reflect the physiologic binding of ligands to their receptors. As reported by Kuhar (48), in vitro procedures have certain advantages over in vivo procedures: the quantity of radioisotope is much smaller than that used in in vivo radioautography and the physiologic condition of the receptor sites can be regulated easily by removing endogenous ligand sources. The present paper mainly deals with the in vivo procedure.
Macroradioautography for histologic receptor distribution
Macroradioautography includes whole body radioautography of animals and organ radioautography of large organs such as the brain, liver and kidney. The procedure for in vivo whole body radioautography is as follows (47,49,50): 1) After intravenous injection of a radiolabeled ligand, the experimental animals are perfused with Ringer solution to wash out unbound ligands from the whole body. Three minutes before the perfusion, the mice are anesthetized by intraperitoneal injection of sodium pentobarbital. 2) The region incised for perfusion is covered with 3% carboxymethylcellulose that has been frozen with powdered dry ice. The entire body of the animal is then frozen at -70oC in a mixture of dry ice and acetone. 3) The frozen animal is embedded in 6% carboxymethylcellulose on the microtome stage and after equilibrating the block with the temperature of the cryostat (-20oC), 20-µm thick whole body cryosections are prepared using a heavy duty microtome (LKB 2250, Sweden). 4) Adhesive tape (Scotch tape, Type 800, 3M Co., St. Paul, MN) is applied to the cut surface of the frozen animal to prevent the section from falling apart. 5) The sections obtained are freeze-dried in a cryostat or deep freezer. The freeze-dried sections are brought to room temperature in a desiccator containing silica gel. 6) The dry sections are then placed in direct contact with films, using aluminum plates that are screwed down. For 125I-labeled ligands, Ultrofilm (LKB, Bethesda, MD), Hyperfilm-3H (Amersham International plc., Buckinghamshire, UK), or a Konica Macroradioautograph (Konica Co., Tokyo, Japan) film should be used (without a protective layer of gelatin), and inserts should be avoided. Before examination using this method of direct contact with the film, negative and positive chemography should be checked (51-53). 7) After exposure in a cool, dark box, the films are developed and fixed. D19 (Kodak, Rochester, NY) is a good emulsion developer because it shows a large range of gray levels and results in good proportionality between absorbance and radioactivity (54). There is an alternative method of whole body radioautography in which whole body cryosections on glass slides are used rather than cryosections adhering to adhesive tape (55).
Whole body radioautographs thus obtained are usually observed with the unaided eye. However, whole body radioautographs are also suitable for observation under a low-power microscope when 125I-labeled ligands are used. To identify organs and tissues appearing in whole body radioautographs, whole body histologic sections that correspond to the radioautographs are prepared (56-58).
With whole body radioautography, radioactivities in various parts of organs and tissues can be estimated and compared. For tubular organs, these techniques are not sufficient for such demands, because a single whole body section does not display any tubular organ as a whole. For this purpose, radioautography of tubular organs has been established (59). For parenchymal organs such as the brain, liver and kidney, the following organ radioautography is recommended: after injection of 125I-labeled ligand, with or without excess unlabeled ligand, the animals are perfused with Ringer solution and fixed with 4% paraformaldehyde solution through the left ventricle. The organs are then removed and immersed in the solution for 2 h. After immersion, the specimens are dehydrated with a graded series of ethanol, and embedded in paraffin after immersion in xylene. Five-µm thick paraffin sections on glass slides are deparaffinized and brought into contact with the film for macroradioautography. After appropriate exposure, the films are developed and fixed. In this case, a nuclear emulsion instead of a film will give better results. The emulsion is applied using the dipping technique (60,61), but the exposure time is much longer than in microradioautography.
In vitro techniques are essential for the study of receptors in tissues and organs with a functional barrier to a given ligand such as the brain. In the in vitro procedure, the organs are removed from the animals without the injection of a radiolabeled ligand. The organs are washed with ice-cold physiologic saline to remove blood and are frozen as soon as possible in isopentane cooled with liquid N2. Sections, 25-µm thick, are cut with a microtome at -15oC and thaw mounted on glass slides. To prevent the section from peeling off the glass slide, the glass slides must be coated with dichlorodimethyl-silane, poly-L-lysine, or gelatin. The sections are dried immediately and preincubated with a buffer solution for 15 to 30 min to remove intrinsic ligands and to act as an inhibitor for peptidase. After preincubation, the sections are incubated with a radiolabeled ligand. Temperature and incubation time are determined by biochemical receptor-binding experiments with homogenate tissue or with the crude membrane fraction. To decrease nonspecific binding, washing with ice-cold isotonic buffer solution is performed after incubation with the radiolabeled ligand. Final washings are conducted with ice-cold distilled H2O to remove salts present in the buffer. After air drying, the sections are exposed to a photographic film.
For analysis of macroradioautographs, the density of the radioautographic images in the films is determined by computer-assisted densitometry. As a standard for quantification of radioautographs, 125I scales ([I-125]micro-scales, RPA522, Amersham International plc.) are used. With the 125I-plastic standards the sensitivity of the film to 125I should be determined (62). To obtain appropriate absorbance values for the radioautographs, it is necessary to know the relationship between isotope concentrations and film blackings at various exposure times. Therefore, in the experiments of radiolabeled ligand binding, with or without unlabeled ligand using serial cryosections, appropriate absorbance values for each section should be obtained by changing the exposure time for the different amount of binding.
Microradioautography for histologic receptor distribution
To determine where specific ligand-binding sites are located at the cellular level, radioautographic techniques with high spatial resolution are required. For this purpose, microradioautography at the light microscopy level is used. The procedure for microradioautography is as follows.
The paraffin- and resin-embedded sections or the frozen sections of tissues obtained from the animals injected with radioactive ligand are used for light microscopic radioautography. These sections are mounted on microscope slides and a nuclear emulsion is applied. For microradioautography several types of nuclear emulsions are commercially available. For microradioautographic studies of receptors at the light microscopy level with 125I, Kodak NTB2, and Konica NR-M2 (Konica Co., Tokyo, Japan) are excellent. Although there are various methods for applying emulsion to a section, the dipping technique is commonly used. The dipping technique is based on brief dipping of the section-slide into a liquefied emulsion, followed by drying of the emulsion. Under safelighting, the appropriate amount of emulsion is transferred from the bottle to a glass cylinder and placed in a water bath at 43oC for 10 min to melt the emulsion. The volume should then be increased by about 50% with distilled water depending on the emulsion used and the thickness of the emulsion layer desired. The molten emulsion is poured into a dipping jar and 1% glycerol is added. A slide is dipped into the emulsion for several seconds and slowly withdrawn. The back of the slide is wiped with paper, and the slide is then cooled quickly by laying it on a metal plate cooled with ice to prevent redistribution of silver grains. After about 10 min on the plate, the slide is laid flat on the bench for 1 h, transferred to a slide box, and the box is kept overnight in a desiccator with silica gel. After appropriate exposure in a cool dark room without silica gel, the emulsion is developed and fixed. The sections are then stained and mounted by routine histologic procedures.
Radioautographs prepared by microradioautography are usually observed by light microscopy. In the radioautographs prepared with a thin layer of emulsion and a 1-µm thick plastic section, it is not only relatively easy to relate silver grains and tissue, but a micrograph can be also taken with both the silver grains and the tissue section in a single focus. However, with thick sections made from paraffin and frozen blocks, using the ordinary dipping method, the resulting distances between the silver grains and tissue might be variable. There are many problems with these stained radioautographs, but they can be solved with a laser scanning microscope (LSM) (63-65).
Microradioautographs are quantified by grain counting. The counting can be carried out on photomicrographs taken by the recorder fitted to the LSM or by image analysis of the photomicrographs. The number of grains in the cells of interest are scored, and the average number of grains per µm2 is determined.
To estimate specific ligand binding among the tissues and cells, statistical analysis is performed, usually by the t-test. In receptor radioautography, however, two different kinds of microradioautographs should be made to identify specific ligand binding. Specific binding of ligand in a given tissue is obtained from the differences between total and nonspecific binding. The total binding is obtained from radioautographs made from the tissue injected with radiolabeled ligand. The nonspecific binding is obtained from those made from the same radiolabeled ligand-injected tissue plus excess unlabeled ligand. Radioautographs showing total binding of a ligand and those showing the nonspecific binding are obtained from different specimens in receptor radioautography. In microradioautography using nuclear emulsions, the grains in a large number of micro-radioautographs are counted, making it difficult to correlate them. For this purpose, a statistical method for receptor radioautography has been developed (66,67).
Histologic distribution of insulin receptors
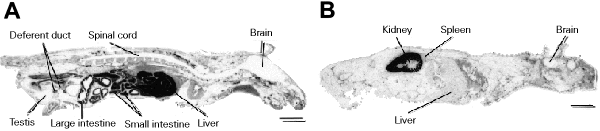
The distribution of insulin receptors in whole animal tissues has been studied in our laboratory by in vivo whole body radioautography (62). After intravenous injection of 125I-insulin into male adult mice, high specific insulin binding occurred in the liver, small intestine and large intestine (Figure 1). Relatively high binding was observed in the pancreas, Harderian gland, and choroid plexus in the brain. The deferent duct also showed relatively high binding, while the binding level in other parts of the reproductive organs, including the testis, was very low. The skeletal muscle and fat, both of which are thought to be major targets for insulin action, showed extremely low insulin binding. Microradioautography of skeletal muscle demonstrated that the blood vessel in the skeletal muscle showed substantial specific insulin binding, but specific binding was not seen in the muscle fiber even after long exposure (Figure 2).
- Whole body radioautographs showing the distribution of total (A) and nonspecific (B) insulin-binding sites in mice 3 min after 125I-insulin injection. The mice were injected intravenously with 185 kBq of 125I-insulin (porcine, 125I-TyrA14 insulin dissolved in Ringer solution at a concentration of 185 kBq/0.4 ml) in the absence (A) or presence (B) of excess (50 µg) unlabeled insulin. Bar = 1 cm.
- Overlay image of differential interference contrast (DIC) and confocal LSM images of microradioautographs of mouse skeletal muscle 3 min after 125I-insulin injection. The number of silver grains on the wall of blood vessels (BV) are higher than that found in skeletal muscle fibers (SMF). Bar = 50 µm.
In the liver, specific insulin receptors on the plasma membrane of hepatocytes have been demonstrated by in vivo radioautographic studies (68,69). The distribution of insulin-binding sites in the liver of fed and fasted mice was studied by microradioautography 3 min after intravenous injection of 125I-insulin (70). Specific binding of 125I-insulin to liver parenchymal cells was seen in these mice. In both fed and fasted mice, a density gradient of the binding from the periportal zone to the perivenous zone was evident, and binding in each zone was significantly higher in fasted than in fed mice.
The presence of insulin receptors in the epithelial cells of the gastrointestinal tract of the rat has been shown by in vivo radioautography (71). Macroradioautographic study of the alimentary tract of mice injected intravenously with 125I-insulin demonstrated a gradual decrease in insulin binding from the duodenum to the rectum (72). This finding is in good agreement with the in vivo radioreceptor assay data of Whitcomb et al. (73). Our in vivo whole body radioautography also demonstrated the absence of insulin-binding sites on the epithelial cells in the non-glandular part of the stomach (62).
As to the pancreas, in vivo microscopic radioautography with 125I-insulin revealed that the exocrine pancreatic cells of the rat have a large number of insulin receptors (71,74). The existence of insulin receptors not only on the plasma membrane of the exocrine pancreatic cells but also on that of duct cells was confirmed by in vivo radioautography (75).
Insulin has been detected in the brain (7,8,76,77), which was thought to be an insulin-independent organ because insulin cannot pass through the blood-brain barrier. The distribution of insulin receptors was investigated in the brain by in vitro radioautography (78-81), and insulin receptor mRNA was demonstrated in rat brain by in situ hybridization (82). These studies demonstrated that the distribution of insulin receptor-binding sites was consistent with the distribution of insulin receptor mRNA, and insulin receptors were most abundant in the granule cell layers of the olfactory bulb, cerebellum and dentate gyrus, in the pyramidal cell layers of the pyriform cortex and hippocampus, in the choroid plexus and in the arcuate nucleus of the hypothalamus. Our detailed studies on the anatomical distribution of insulin receptors in the mouse hippocampus using radioautography after in vitro labeling of cryostat sections with 125I-insulin demonstrated that insulin receptors were distributed most intensely in the granular and pyramidal layers, while the densities of insulin receptors were low in the lacuno-molecular layer (83). Among the pyramidal cell layers of the hippocampus, CA3b and CA3c sectors showed significantly higher densities of insulin receptors than CA1 and CA3a. Baskin et al. (78), studying rat brain by quantitative radioautography, showed that the choroid plexus had a high density of insulin receptors.
The kidney showed an intense radioautographic reaction when experimental animals received radiolabeled insulin, but the binding of labeled insulin was nonspecific in nature since the strong reaction was not depressed by the presence of an excess amount of unlabeled insulin (69,84). In vitro radioautographic study, however, demonstrated specific insulin receptors in the glomeruli and tubules of the cortex (85).
In the reproductive system, the deferent duct showed relatively high insulin binding in the in vivo whole body autoradiograph (62). In vivo microradioautographic analysis of insulin-binding sites in the mouse deferent duct (86) showed the presence of specific insulin binding in the endothelial cells of capillaries and certain fibroblasts in the lamina propria, but the epithelial cells, except for the basal cells, did not show any insulin binding (Figure 3). Endothelial cells are known to be insulin targets (87), and the presence of specific insulin receptors on the vascular endothelial cells has been demonstrated by in vivo radioautography in rat heart capillary (88). It has been shown that insulin has a biological effect on cultured human lung fibroblasts (89), probably mediated by an interaction of insulin with insulin-like growth factor I receptor. However, although insulin binding to fibroblasts in the mouse deferent duct decreased significantly in the presence of an excess amount of insulin-like growth factor I (86), the rate of decrease was 22% indicating that, to some extent, the receptors are specific for insulin. Hirose et al. (86) also indicated that the fibroblasts in the deferent duct do not always show insulin binding and two types of fibroblasts could be distinguished by electron microscopy. The other reproductive organs such as testis, prostate, seminal vesicle, and epididymis have been shown to possess insulin receptors by the membrane binding assay (90,91). A radioautographic localization study demonstrated the presence of insulin receptors in rat Leydig cells (92).
- Overlay image of differential interference contrast (DIC) and confocal LSM images of microradioautographs of mouse deferent duct 3 min after intravenous injection of 125I-insulin. Numerous silver grains can be seen in the lamina propria (LP), but not in the epithelium (ET) or smooth muscle layer (SML). Bar = 50 µm.
In skeletal and smooth muscles, in vivo radioautographic investigation failed to demonstrate specific insulin binding (62,71), though the muscles are clearly major target organs of insulin and many biochemical studies on muscle insulin receptors have been performed (93,94). The situation of the adipocytes is equal to that of muscle cells. The reason why we have not been able to demonstrate insulin binding in these tissues by in vivo radioautography is uncertain at present.
Apart from the tissues described above, localization of insulin receptors has been demonstrated by in vivo radioautography in osteoblasts of rat tibia (84), parenchymal cells of the adrenal cortex and medulla (71), and epidermal cells of mouse skin (95). Immunohistochemical localization of insulin receptors has been demonstrated in the syncytiotrophoblast of human placenta from 6 to 10 weeks postmenstruation (96), in human fetal fibroblasts (97), and in amacrine cells of the chick retina (98).
Histologic distribution of glucagon receptors
The receptors for glucagon have been identified in the kidney (99,100), brain (101,102), lymphoid cells of the spleen and thymus (103), parenchymal cells of the liver (104-106), and endothelial and Kupffer cells in the liver (107), heart (108-110), adipose tissue (100), intestinal smooth muscle tissue (33) and endocrine pancreatic cells (111,112). Recently, expression of glucagon receptor mRNA was also examined in various tissues revealing that liver, kidney, heart, adipose tissue, spleen, pancreatic islets, ovary, and thymus expressed relatively abundant levels of glucagon receptor mRNA, whereas levels in the stomach, small intestine, adrenal glands, thyroid and skeletal muscle were low (113). Similar results have been reported by Svoboda et al. (114), Burcelin et al. (115), Christophe (116), and Yoo-Warren et al. (117). However, expression of glucagon receptor mRNA does not necessarily signify the formation of glucagon receptor, because it has been suggested that glucagon receptor expression is modulated at a step after mRNA formation (117).
Only limited information is available concerning the histologic distribution of glucagon receptors in vivo in liver parenchymal cells (105,106), intestinal smooth muscle cells (33) and the brain (102). We have investigated the histologic distribution of the receptors in young, adult and pregnant mice using whole body and light microscopic radioautography with 125I-labeled glucagon.
The whole body radioautographic experiment using adult mice injected intravenously with 125I-glucagon demonstrated that only the liver had very high glucagon binding (Figure 4). In vivo microradioautography of the liver revealed the presence of a density gradient of binding from the periportal zone to the perivenous zone (Figure 5). Some cortical tissues in the kidney also showed significant specific binding, but the level was much lower than in the liver. In other tissues and organs, specific binding was equal to or below the reliable quantification limit.
- Whole body radioautographs showing the distribution of total (A) and nonspecific (B) glucagon-binding sites in mice 3 min after intravenous injection of 185 kBq 125I-glucagon. 125I-glucagon (3-[125I]iodotyrosyl10glucagon dissolved in Ringer solution at a concentration of 185 kBq/0.4 ml) was injected into the tail vein in the absence (A) or presence (B) of excess (25 µg) unlabeled glucagon. Bar = 1 cm.
- Overlay images of differential interference contrast (DIC) and confocal LSM images of microradioautographs of mouse liver 3 min after intravenous injection of 125I-glucagon. The number of silver grains in the periportal zone (A) is higher than that in the perivenous zone (B). PV, Portal vein; CV, central vein. Bar = 50 µm.
Conclusion
A working hypothesis of a direct correlation of hormone receptor density with hormone action points to hitherto unemphasized targets in the small and large intestines and deferent duct as major sites of insulin action in the body. In contrast, only the liver is regarded as a major site of glucagon action. However, the existence of insulin receptors has been demonstrated in almost all tissues studied. Furthermore, certain tissues such as skeletal muscle and adipose tissue revealed the existence of insulin receptors despite the difficulty of morphological demonstration of insulin receptors in these tissues. These conflicts may derive from the sensitivity of the technique used for detecting the hormone receptors. The function of the hormone receptor in a given tissue should change with age (93) and the action of a given hormone should change in accordance with concentration (118). In addition to these factors, evidence has pointed to heterogeneity of insulin receptor structure and function (119). To understand the true function of insulin and glucagon in a specific tissue, anatomical localization of the receptors should be the first step of investigation.
1. Cuello AC (1983). Brain Microdissection Techniques. John Wiley & Sons, Chichester.
2. MacLusky NJ, Brown TJ, Jones E, Leranth C & Hochberg RB (1990). Autoradiographic and microchemical methods for quantification of steroid receptors. In: Conn PM (Editor), Quantitative and Qualitative Microscopy. Academic Press, New York, 3-35.
3. Yip CC (1985). Insulin structure: receptor binding and probing structure with photoreactive derivatives. In: Hollenberg MD (Editor), Insulin: its Receptor and Diabetes. Marcel Dekker, New York, 21-37.
4. Felig P & Bergman M (1995). The endocrine pancreas: Diabetes mellitus. In: Felig P, Baxter JD & Frohman LA (Editors), Endocrinology and Metabolism. 3rd edn. McGraw-Hill, New York, 1107-1250.
5. Kahn CR & Goldfine AB (1993). Molecular determinants of insulin action. Journal of Diabetes and its Complications, 7: 92-105.
6. Gammeltoft S (1984). Insulin receptors: binding kinetics and structure-function relationship of insulin. Physiological Reviews, 64: 1321-1378.
7. Havrankova J, Roth J & Brownstein M (1978). Insulin receptors are widely distributed in the central nervous system of the rat. Nature, 272: 827-829.
8. Havrankova J, Schmechel D, Roth J & Brownstein M (1978). Identification of insulin in rat brain. Proceedings of the National Academy of Sciences, USA, 75: 5737-5741.
9. De Meyts P, Roth J, Neville Jr DM, Gavin III JR & Lesniak MA (1973). Insulin interactions with its receptors: experimental evidence for negative cooperativity. Biochemical and Biophysical Research Communications, 55: 154-161.
10. Jacobs S (1985). Insulin receptor: purification and structure. In: Hollenberg MD (Editor), Insulin: its Receptor and Diabetes. Marcel Dekker, New York, 39-55.
11. Seino S, Seino M, Nishi S & Bell GI (1989). Structure of the human insulin receptor gene and characterization of its promoter. Proceedings of the National Academy of Sciences, USA, 86: 114-118.
12. Seino S & Bell GI (1989). Alternative splicing of human insulin receptor messenger RNA. Biochemical and Biophysical Research Communications, 159: 312-316.
13. Benecke H, Flier JS & Moller DE (1992). Alternatively spliced variants of the insulin receptor protein. Expression in normal and diabetic human tissues. Journal of Clinical Investigation, 89: 2066-2070.
14. McClain DA (1991). Different ligand affinities of the two human insulin receptor splice variants are reflected in parallel changes in sensitivity for insulin action. Molecular Endocrinology, 5: 734-739.
15. Yamaguchi Y, Flier JS, Yokota A, Benecke H, Backer JM & Moller DE (1991). Functional properties of two naturally occurring isoforms of the human insulin receptor in Chinese hamster ovary cells. Endocrinology, 129: 2058-2066.
16. Lee J & Pilch PF (1994). The insulin receptor: structure, function, and signaling. American Journal of Physiology, 266: C319-C334.
17. Lienhard GE (1994). Life without the IRS. Nature, 372: 128-129.
18. Lee YC, Brubaker PL & Drucker DJ (1990). Developmental and tissue-specific regulation of proglucagon gene expression. Endocrinology, 127: 2217-2222.
19. Lee YC, Campos RV & Drucker DJ (1993). Region- and age-specific differences in proglucagon gene expression in the central nervous system of wild-type and glucagon-simian virus-40 T-antigen transgenic mice. Endocrinology, 133: 171-177.
20. Agungpiryono S, Yamada J, Kitamura N, Yamamoto Y, Said N, Sigit K & Yamashita T (1994). Immunohistochemical study of the distribution of endocrine cells in the gastrointestinal tract of the lesser mouse deer (Tragulus javanicus). Acta Anatomica, 151: 232-238.
21. Alison BC (1990). The ontogeny and distribution of glucagon- and pancreatic polypeptide-immunoreactive cells in the gastrointestinal tract of the chicken. Anatomy and Embryology, 182: 605-610.
22. Sjolund K, Sanden G, Hakanson R & Sundler F (1983). Endocrine cells in human intestine: an immunocytochemical study. Gastroenterology, 85: 1120-1130.
23. Jin SL, Han VK, Simmons G, Towle AC, Lauder JM & Lund PK (1988). Distribution of glucagon-like peptide I (GLP-I), glucagon, and glicentin in the rat brain: an immunocytochemical study. Journal of Comparative Neurology, 271: 519-532.
24. Under HR & Orci L (1995). Glucagon secretion, alpha cell metabolism, and glucagon action. In: Degree LJ (Editor), Endocrinology. 3rd edn. WB Saunders, Philadelphia, 1337-1353.
25. Mojsov S, Heinrich G, Wilson IB, Ravazzola M, Orci L & Habener JF (1986). Preproglucagon gene expression in pancreas and intestine diversifies at the level of post-translational processing. Journal of Biological Chemistry, 261: 11880-11889.
26. Unson CG, Wu CR & Merrifield RB (1994). Roles of aspartic acid 15 and 21 in glucagon action: receptor anchor and surrogates for aspartic acid 9. Biochemistry, 33: 6884-6887.
27. Unson CG, Wu CR, Fitzpatrick KJ & Merrifield RB (1994). Multiple-site replacement analogs of glucagon. A molecular basis for antagonist design. Journal of Biological Chemistry, 269: 12548-12551.
28. Hagen JH (1961). Effect of glucagon on the metabolism of adipose tissue. Journal of Biological Chemistry, 236: 1023-1027.
29. Burns TW & Langley P (1968). Observations on lipolysis with isolated adipose tissue cells. Journal of Laboratory and Clinical Medicine, 72: 813-823.
30. Hansen MD, Heiling VJ & Miles JM (1991). Effects of glucagon on free fatty acid metabolism in humans. Journal of Clinical Endocrinology and Metabolism, 78: 308-315.
31. Gregersen H, Ritting S, Kraglund K & Phillips S (1988). The effect of glucagon and glucagon-(1-21)-peptide on antroduodenal motility in healthy volunteers. Scandinavian Journal of Gastroenterology, 152: 42-47.
32. MacLeod KM, Rodgers RL & McNeill JH (1981). Characterization of glucagon-induced changes in rate, contractility and cyclic AMP levels in isolated cardiac preparations of the rat and guinea pig. Journal of Pharmacology and Experimental Therapeutics, 217: 798-804.
33. Santamaria L, De Miguel E, Codesal J, Ramirez JR & Picazo J (1991). Identification of glucagon binding sites on smooth muscle tissue of dog intestine; quantification by means of ultrastructural autoradiography. Anatomischer Anzeiger, 172: 149-157.
34. Van Schravendijk CFH, Foriers A, Hooghe-Peters EL, Rogiers V, De Meyts P, Sodoyez JC & Pipeleers DG (1985). Pancreatic hormone receptors on islet cells. Endocrinology, 117: 841-848.
35. Kofod H, Unson C & Merrifield RB (1988). Potentiation of glucose-induced insulin release in islets by desHis1[Glu9]glucagon amide. International Journal of Peptide and Protein Research, 32: 436-440.
36. Abrahamsen N & Nishimura E (1995). Regulation of glucagon and glucagon-like peptide-1 receptor messenger ribonucleic acid expression in cultured rat pancreatic islets by glucose, cyclic adenosine 3',5'-monophosphate, and glucocorticoids. Endocrinology, 136: 1572-1578.
37. Bailly C, Imbert-Teboul M, Chabardés D, Hus-Citharel A, Montégut M, Clique A & Morel F (1980). The distal nephron of rat kidney: A target site for glucagon. Proceedings of the National Academy of Sciences, USA, 77: 3422-3424.
38. Delahouse M, Mercier O, Bichara M, Paillard M, Leviel F & Assan R (1988). Glucagon inhibits urinary acidification in the rat. American Journal of Physiology, 254: F762-F769.
39. Lok S, Kuijper JL, Jelinek LJ, Kramer JM, Whitmore TE, Sprecher CA, Mathewes S, Grant FJ, Biggs SH, Rosenberg GB, Sheppard PO, O'Hara PJ, Foster D & Kindsvogel W (1994). The human glucagon receptor encoding gene: structure, cDNA sequence and chromosomal localization. Gene, 140: 203-209.
40. Jelinek LJ, Lok S, Rosenberg GB, Smith RA, Grant FJ, Biggs S, Bensch PA, Kuijper JL, Sheppard PO, Sprecher CA, O'Hara PJ, Foster D, Walker KM, Chen LHJ, McKernan PA & Kindsvogel W (1993). Expression cloning and signaling properties of the rat glucagon receptor. Science, 259: 1614-1616.
41. Svoboda M, Ciccarelli E, Tastenoy M, Robberecht P & Christophe J (1993). A cDNA construct allowing the expression of rat hepatic glucagon receptors. Biochemical and Biophysical Research Communications, 192: 135-142.
42. Unson CG, Cypess AM, Kim HN, Goldsmith PK, Carruthers CJ, Merrifield RB & Sakmar TP (1995). Characterization of deletion and truncation mutants of the rat glucagon receptor. Seven transmembrane segments are necessary for receptor transport to the plasma membrane and glucagon binding. Journal of Biological Chemistry, 270: 27720-27727.
43. MacNeil DJ, Occi JL, Hey PJ, Strader CD & Graziano MP (1994). Cloning and expression of a human glucagon receptor. Biochemical and Biophysical Research Communications, 198: 328-334.
44. Wakelam MJ, Murphy GJ, Hruby VJ & Houslay MD (1986). Activation of two signal-transduction systems in hepatocytes by glucagon. Nature, 323: 68-71.
45. Iyengar R & Herberg JT (1984). Structural analysis of the hepatic glucagon receptor. Identification of a guanine nucleotide-sensitive hormone-binding region. Journal of Biological Chemistry, 259: 5222-5229.
46. Padrell E, Herberg JT, Monsatirsky B, Floyd G, Premont RT & Iyengar R (1987). The hepatic glucagon receptor: a comparative study of the regulatory and structural properties. Endocrinology, 120: 2316-2325.
47. Shimada M & Watanabe M (1993). The techniques and applications of whole-body radioautography. In: Nagata T (Editor), Radioautography in Medicine. Shinshu University Press, Matsumoto, Japan, 33-40.
48. Kuhar MJ (1991). Perspectives on receptor autoradiography in human brain. In: Mendelsohn FAD & Paxinos G (Editors), Receptors in the Human Nervous System. Academic Press, San Diego.
49. Curtis CG, Cross SAM, McCulloch RJ & Powell GM (1981). Whole-Body Autoradiography. Academic Press, London.
50. Shimada M & Watanabe M (1995). Recent progress in whole-body radioautography. Cellular and Molecular Biology, 41: 39-48.
51. Rogers AW & Watanabe M (1988). The relative sensitivities of autoradiographic emulsions to negative chemography. Journal of Microscopy, 149: 193-197.
52. Watanabe M & Rogers AW (1987). Prevention of negative chemography in phenylenediamine stained autoradiographs of plastic semithin sections. Stain Technology, 62: 173-177.
53. Watanabe M, Koide T, Hayasaki H, Kanbara K, Jo N & Shimada M (1994). Negative chemography with mouse skeletal muscle. Bulletin of the Osaka Medical College, 40: 17-19.
54. Segu L, Rage P, Pinard R & Boulenguez P (1992). Computer-assisted quantitative receptor autoradiography. In: Conn PM (Editor), Computers and Computations in the Neurosciences. Academic Press, San Diego, 129-143.
55. Watanabe M, Hirose Y & Shimada M (1991). A new technique for autoradiography of whole-body sections mounted on glass slides. Acta Histochemica et Cytochemica, 24: 167-171.
56. Shimono R, Watanabe M & Goto H (1982). A new method of preparation of whole-body sections for autoradiography and staining. Kaibogaku Zasshi, 57: 15-21.
57. Watanabe M, Kihara T, Shimada M & Kurimoto K (1978). Preparation and staining of whole-body sections. Cellular and Molecular Biology, 23: 311-315.
58. Watanabe M, Shimada M & Kurimoto K (1975). Staining method for whole-body autoradiography. Stain Technology, 50: 239-243.
59. Kuno N, Watanabe M, Akagi N, Sugimoto M & Kihara T (1987). A technique of autoradiography for tubular organs. Acta Histochemica et Cytochemica, 20: 547-555.
60. Rogers AW (1979). Techniques of Autoradiography. 3rd edn. Elsevier North-Holland, Amsterdam.
61. Rogers AW (1979). Practical Autoradiography. Review No. 20. Amersham International Limited, Amersham, UK.
62. Watanabe M, Hirose Y, Sugimoto M, Nakanishi M, Watanabe H & Shimada M (1992). The distribution of tissue insulin receptors in the mouse by whole-body autoradiography. Journal of Receptor Research, 12: 13-37.
63. Watanabe M, Kawamoto S, Nakatsuka Y, Koide T & Shimada M (1993). An application of the laser scanning microscope to microautoradiography. Acta Histochemica et Cytochemica, 26: 93-99.
64. Watanabe M, Koide T, Konishi M, Kanbara K & Shimada M (1995). Use of confocal laser scanning microscopy in radioautographic study. Cellular and Molecular Biology, 41: 131-136.
65. Watanabe M, Koide T, Nakatsuka Y & Shimada M (1993). Visualization of microradioautography by confocal laser scanning microscope. In: Nagata T (Editor), Radioautography in Medicine. Shinshu University Press, Matsumoto, Japan, 215-219.
66. Watanabe M, Nishimura Y, Jo N, Takafuchi M, Kiyokane K & Shimada M (1996). Statistical analysis for receptor autoradiography. Acta Histochemica et Cytochemica, 29: 135-139.
67. Nishimura Y, Watanabe M, Jo N & Shimada M (1997). The difference of means of two indirect unpaired groups in receptor autoradiography. Journal of Biopharmaceutical Statistics, 7: 441-452.
68. Bergeron JJM, Levine G, Sikstrom R, O'Shaughnessy D, Kopriwa B, Nadler NJ & Posner BI (1977). Polypeptide hormone binding sites in vivo. Initial localization of 125I-labeled insulin to hepatocyte plasma membrane as visualized by electron microscope radioautography. Proceedings of the National Academy of Sciences, USA, 74: 5051-5055.
69. Bergeron JJM, Sikstrom R, Hand AR & Posner BI (1979). Binding and uptake of 125I-insulin into rat liver hepatocytes and endothelium. An in vivo radioautographic study. Journal of Cell Biology, 80: 427-443.
70. Nakanishi M, Watanabe M & Shimada M (1995). Heterogeneous distribution of 125I-insulin binding sites in the liver of fed and fasted mice. Cellular and Molecular Biology, 41: 137-144.
71. Bergeron JJM, Rachubinski R, Searle N, Borts D, Sikstrom R & Posner BI (1980). Polypeptide hormone receptors in vivo: Demonstration of insulin binding to adrenal gland and gastrointestinal epithelium by quantitative radioautography. Journal of Histochemistry and Cytochemistry, 28: 824-835.
72. Shimada M & Watanabe M (1997). Whole-body and microradioautography of receptors. In: Morel G (Editor), Visualization of Receptors: Methods in Light and Electron Microscopy. CRC Press, Paris, 97-124.
73. Whitcomb DC, O'Dorison TM, Cataland S, Shetzline MA & Nishikawara MT (1985). Identification of tissue insulin receptors: use of a unique in vivo radioreceptor assay. American Journal of Physiology, 249: E561-E567.
74. Bergeron JJM, Rachubinski R, Searle N, Sikstrom R, Borts D, Bastian P & Posner BI (1980). Radioautographic visualization of in vivo insulin binding to the exocrine pancreas. Endocrinology, 107: 1069-1080.
75. Sakamoto C, Williams JA, Roach E & Goldfine ID (1984). In vivo localization of insulin binding to cells of the rat pancreas. Proceedings of the Society for Experimental Biology and Medicine, 175: 497-502.
76. Dorn A, Bernstein HG, Hahn AJ, Zeigler M & Rummelfanger H (1981). Insulin immunohistochemistry of rodent CNS: apparent species differences but good correlation with radioimmunochemical data. Histochemistry, 71: 609-616.
77. Pansky B & Hatfield JS (1978). Cerebral localization of insulin by immunofluorescence. American Journal of Anatomy, 153: 459-467.
78. Baskin DG, Brewitt B, Davidson DA, Corp E, Paquette T, Figlewicz DP, Lewellen T, Graham MK, Woods SG & Dorsa DM (1986). Quantitative autoradiographic evidence for insulin receptors in the choroid plexus of the rat brain. Diabetes, 35: 246-249.
79. Baskin DG, Davidson DA, Corp E, Lewellen T & Graham M (1986). An inexpensive microcomputer digital imaging system for densitometry: quantitative autoradiography of insulin receptors with 125I and LKB Ultrofilm. Journal of Neuroscience Methods, 16: 119-129.
80. Hill JM, Lesniak MA, Pear CB & Roth J (1986). Autoradiographic localization of insulin receptors in rat brain: prominence in olfactory and limbic areas. Neuroscience, 17: 1127-1138.
81. Werther GA, Hogg A, Oldfield BJ, McKinley MJ, Figdor R, Allen M & Mendelsohn FAO (1987). Localization and characterization of insulin receptors in rat brain and pituitary gland using in vitro autoradiography and computerized densitometry. Endocrinology, 121: 1562-1570.
82. Marks JL, Porte Jr D, Stahl WL & Baskin DG (1990). Localization of insulin receptor mRNA in rat brain by in situ hybridization. Endocrinology, 127: 3234-3236.
83. Shimada M, Hirose Y, Nakanishi M, Watanabe H, Kawamoto S & Watanabe M (1992). Autoradiographic studies on insulin receptor of the mouse hippocampus. In: Wegmann RJ & Wegmann MA (Editors), Recent Advances in Cellular and Molecular Biology. Vol. 4. Peeters Press, Leuven, Belgium, 271-278.
84. Martineau Doizé B, Warshawsky MH & Bergeron JJM (1986). In vivo demonstration by radioautography of binding sites for insulin in liver, kidney, and calcified tissues of the rat. Anatomical Record, 214: 130-140.
85. Sechi LA, De Carli S & Bartoli E (1994). In situ characterization of renal insulin receptors in the rat. Journal of Receptor Research, 14: 347-356.
86. Hirose Y, Watanabe M, Koide T & Shimada M (1994). Insulin-binding sites in the mouse deferent duct. Acta Histochemica, 96: 405-414.
87. Vinters HV & Berliner JA (1987). The blood vessel wall as an insulin target tissue. Diabete et Metabolisme, 13: 294-300.
88. Bar RS, Boes M & Sandra A (1988). Vascular transport of insulin to rat cardiac muscle. Central role of the capillary endothelium. Journal of Clinical Investigation, 81: 1225-1233.
89. Goldstein RH, Poliks CF, Pilch PF, Smith BD & Fine A (1989). Stimulation of collagen formation by insulin and insulin-like growth factor I in cultures of human lung fibroblasts. Endocrinology, 124: 964-970.
90. Loubassou S, Grizard G & Grizard J (1989). Insulin binding sites in solubilized membranes from rat testis. Effect of age. Andrologia, 21: 377-383.
91. Saucier J, Dube JY & Tremblay RR (1981). Specific insulin binding sites in rat testis: characterization and variation. Endocrinology, 109: 2220-2225.
92. Abele V, Pelletier G & Tremblay RR (1986). Radioautographic localization and regulation of the insulin receptors in rat testis. Journal of Receptor Research, 6: 461-473.
93. Carvalho CR, Brenelli SL, Silva AC, Nunes AL, Velloso LA & Saad MJ (1996). Effect of aging on insulin receptor, insulin receptor substrate-1, and phosphatidylinositol 3-kinase in liver and skeletal muscle. Endocrinology, 137: 151-159.
94. Munoz P, Rosemblatt M, Testar X, Palacin M, Thoidid G, Pilch PF & Zorzano A (1995). The T-tubule is a cell surface target for insulin-regulated recycling of membrane proteins in skeletal muscle. Biochemical Journal, 312: 393-400.
95. Jo N, Watanabe M, Kiyokane K & Shimada M (1997). In vivo microradioautographic study of insulin binding in the skin of normal and NIDDM mice: with special reference to acanthosis nigricans. Cellular and Molecular Biology, 43: 157-164.
96. Desoye G, Hartmann M, Blaschitz A, Dohr G, Hahn T, Kohnen G & Kaufmann P (1994). Insulin receptors in syncytiotrophoblast and fetal endothelium of human placenta. Immunohistochemical evidence for developmental changes in distribution pattern. Histochemistry, 101: 277-285.
97. Wolf HJ & Desoye G (1993). Immunohistochemical localization of glucose transporters and insulin receptors in human fetal membranes at term. Histochemistry, 100: 379-385.
98. Hyndmann AG (1995). Identification of a population of amacrine cells rich in insulin receptors. Brain Research, Developmental Brain Research, 75: 289-292.
99. Iwanij V (1995). Canine kidney glucagon receptor: evidence for a structurally-different, tissue-specific variant of the glucagon receptor. Molecular and Cellular Endocrinology, 115: 21-28.
100. Iwanij V & Vincent AC (1990). Characterization of the glucagon receptor and its functional domains using monoclonal antibodies. Journal of Biological Chemistry, 265: 21302-21308.
101. Hoosein NM & Gurd RS (1984). Identification of glucagon receptors in rat brain. Proceedings of the National Academy of Sciences, USA, 81: 4368-4372.
102. Montaron A, Moyse E & Barré H (1994). Radioautographic demonstration and localization of glucagon receptors in duck brain. Brain Research, 663: 121-130.
103. Koh WS, Lee M, Yang KH & Kaminski NE (1996). Expression of functional glucagon receptors on lymphoid cells. Life Sciences, 58: 741-751.
104. Frandsen EK & Bacchus RA (1987). Glucagon receptor binding, dissociation and degradation in rat liver plasma membranes studied by a microperfusion method. Biochimica et Biophysica Acta, 929: 74-80.
105. Berthould VM, Iwanij V, Garcia AM & Saez JC (1992). Connexins and glucagon receptors during development of rat hepatic acinus. American Journal of Physiology, 263: G650-G658.
106. Watanabe J, Kanamura S, Asada-Kubota M, Kanai K & Oka M (1984). Receptor-mediated endocytosis of glucagon in isolated mouse hepatocytes. Anatomical Record, 210: 557-567.
107. Watanabe J, Kanai K & Kanamura S (1988). Glucagon receptors in endothelial and Kupffer cells of mouse liver. Journal of Histochemistry and Cytochemistry, 36: 1081-1089.
108. Clark Jr CM, Beatty B & Allen DO (1973). Evidence for delayed development of the glucagon receptor of adenylate cyclase in the fetal and neonatal rat heart. Journal of Clinical Investigation, 52: 1018-1025.
109. Robberect P, Damien C, Moroder L, Coy DH, Wunsch E & Christophe J (1988). Receptor occupancy and adenylate cyclase activation in rat liver and heart membranes by 10 glucagon analogs modified in position 2, 3, 4, 25, 27 and/or 29. Regulatory Peptides, 21: 117-128.
110. Yount EA, Clark JF & Clark Jr CM (1976). Development of guanylylimidodiphosphate-dependent activation of adenylate cyclase by glucagon in the neonatal rat heart. Pediatric Research, 10: 851-854.
111. Desbuquois B & Authier F (1989). Glucagon receptors. Annales d' Endocrinologie, 50: 440-446.
112. Kawai K, Yokota C, Ohashi S, Watanabe Y & Yamashita K (1995). Evidence that glucagon stimulates insulin secretion through its own receptor in rats. Diabetologia, 38: 274-276.
113. Hansen LH, Abrahamsen N & Nishimura E (1995). Glucagon receptor mRNA distribution in rat tissues. Peptides, 16: 1163-1166.
114. Svoboda M, Tastenoy M, Vertongen P & Robberecht P (1994). Relative quantitative analysis of glucagon receptor mRNA in rat tissues. Molecular and Cellular Endocrinology, 105: 132-137.
115. Burcelin R, Li J & Charron MJ (1995). Cloning and sequence analysis of the murine glucagon receptor-encoding gene. Gene, 164: 305-310.
116. Christophe J (1994). Molecular cloning and functional expression of glucagon receptors in the liver, the heart and various other tissues. Bulletin et Memoires de l' Academie Royale de Medecine de Belgique, 149: 271-277.
117. Yoo-Warren H, Willse AG, Hancock N, Hull J, McCaleb M & Livingston JN (1994). Regulation of rat glucagon receptor expression. Biochemical and Biophysical Research Communications, 205: 347-353.
118. Camps M, Guma A, Vinals F, Testar X, Palacin M & Zorzano A (1992). Evidence for the lack of spare high-affinity insulin receptors in skeletal muscle. Biochemical Journal, 285: 993-999.
119. Joost HG (1995). Structural and functional heterogeneity of insulin receptors. Cell Signal, 7: 85-91.
Address for correspondence: M. Watanabe, Department of Anatomy, Osaka Medical College, Takatsuki, Osaka 569, Japan. Fax: 81-726-84-6512. E-mail: an2002@art.osaka-med.ac.jp
Presented at the 5th International Symposium on Radioautography, São Paulo, SP, Brasil, August 24-26, 1997. Received July 11, 1997. Accepted July 22, 1997.
- 2. MacLusky NJ, Brown TJ, Jones E, Leranth C & Hochberg RB (1990). Autoradiographic and microchemical methods for quantification of steroid receptors. In: Conn PM (Editor), Quantitative and Qualitative Microscopy Academic Press, New York, 3-35.
- 3. Yip CC (1985). Insulin structure: receptor binding and probing structure with photoreactive derivatives. In: Hollenberg MD (Editor), Insulin: its Receptor and Diabetes Marcel Dekker, New York, 21-37.
- 4. Felig P & Bergman M (1995). The endocrine pancreas: Diabetes mellitus. In: Felig P, Baxter JD & Frohman LA (Editors), Endocrinology and Metabolism 3rd edn. McGraw-Hill, New York, 1107-1250.
- 5. Kahn CR & Goldfine AB (1993). Molecular determinants of insulin action. Journal of Diabetes and its Complications, 7: 92-105.
- 6. Gammeltoft S (1984). Insulin receptors: binding kinetics and structure-function relationship of insulin. Physiological Reviews, 64: 1321-1378.
- 7. Havrankova J, Roth J & Brownstein M (1978). Insulin receptors are widely distributed in the central nervous system of the rat. Nature, 272: 827-829.
- 8. Havrankova J, Schmechel D, Roth J & Brownstein M (1978). Identification of insulin in rat brain. Proceedings of the National Academy of Sciences, USA, 75: 5737-5741.
- 9. De Meyts P, Roth J, Neville Jr DM, Gavin III JR & Lesniak MA (1973). Insulin interactions with its receptors: experimental evidence for negative cooperativity. Biochemical and Biophysical Research Communications, 55: 154-161.
- 10. Jacobs S (1985). Insulin receptor: purification and structure. In: Hollenberg MD (Editor), Insulin: its Receptor and Diabetes Marcel Dekker, New York, 39-55.
- 11. Seino S, Seino M, Nishi S & Bell GI (1989). Structure of the human insulin receptor gene and characterization of its promoter. Proceedings of the National Academy of Sciences, USA, 86: 114-118.
- 12. Seino S & Bell GI (1989). Alternative splicing of human insulin receptor messenger RNA. Biochemical and Biophysical Research Communications, 159: 312-316.
- 13. Benecke H, Flier JS & Moller DE (1992). Alternatively spliced variants of the insulin receptor protein. Expression in normal and diabetic human tissues. Journal of Clinical Investigation, 89: 2066-2070.
- 14. McClain DA (1991). Different ligand affinities of the two human insulin receptor splice variants are reflected in parallel changes in sensitivity for insulin action. Molecular Endocrinology, 5: 734-739.
- 15. Yamaguchi Y, Flier JS, Yokota A, Benecke H, Backer JM & Moller DE (1991). Functional properties of two naturally occurring isoforms of the human insulin receptor in Chinese hamster ovary cells. Endocrinology, 129: 2058-2066.
- 16. Lee J & Pilch PF (1994). The insulin receptor: structure, function, and signaling. American Journal of Physiology, 266: C319-C334.
- 17. Lienhard GE (1994). Life without the IRS. Nature, 372: 128-129.
- 18. Lee YC, Brubaker PL & Drucker DJ (1990). Developmental and tissue-specific regulation of proglucagon gene expression. Endocrinology, 127: 2217-2222.
- 19. Lee YC, Campos RV & Drucker DJ (1993). Region- and age-specific differences in proglucagon gene expression in the central nervous system of wild-type and glucagon-simian virus-40 T-antigen transgenic mice. Endocrinology, 133: 171-177.
- 20. Agungpiryono S, Yamada J, Kitamura N, Yamamoto Y, Said N, Sigit K & Yamashita T (1994). Immunohistochemical study of the distribution of endocrine cells in the gastrointestinal tract of the lesser mouse deer (Tragulus javanicus). Acta Anatomica, 151: 232-238.
- 21. Alison BC (1990). The ontogeny and distribution of glucagon- and pancreatic polypeptide-immunoreactive cells in the gastrointestinal tract of the chicken. Anatomy and Embryology, 182: 605-610.
- 22. Sjolund K, Sanden G, Hakanson R & Sundler F (1983). Endocrine cells in human intestine: an immunocytochemical study. Gastroenterology, 85: 1120-1130.
- 23. Jin SL, Han VK, Simmons G, Towle AC, Lauder JM & Lund PK (1988). Distribution of glucagon-like peptide I (GLP-I), glucagon, and glicentin in the rat brain: an immunocytochemical study. Journal of Comparative Neurology, 271: 519-532.
- 24. Under HR & Orci L (1995). Glucagon secretion, alpha cell metabolism, and glucagon action. In: Degree LJ (Editor), Endocrinology 3rd edn. WB Saunders, Philadelphia, 1337-1353.
- 25. Mojsov S, Heinrich G, Wilson IB, Ravazzola M, Orci L & Habener JF (1986). Preproglucagon gene expression in pancreas and intestine diversifies at the level of post-translational processing. Journal of Biological Chemistry, 261: 11880-11889.
- 26. Unson CG, Wu CR & Merrifield RB (1994). Roles of aspartic acid 15 and 21 in glucagon action: receptor anchor and surrogates for aspartic acid 9. Biochemistry, 33: 6884-6887.
- 27. Unson CG, Wu CR, Fitzpatrick KJ & Merrifield RB (1994). Multiple-site replacement analogs of glucagon. A molecular basis for antagonist design. Journal of Biological Chemistry, 269: 12548-12551.
- 28. Hagen JH (1961). Effect of glucagon on the metabolism of adipose tissue. Journal of Biological Chemistry, 236: 1023-1027.
- 29. Burns TW & Langley P (1968). Observations on lipolysis with isolated adipose tissue cells. Journal of Laboratory and Clinical Medicine, 72: 813-823.
- 30. Hansen MD, Heiling VJ & Miles JM (1991). Effects of glucagon on free fatty acid metabolism in humans. Journal of Clinical Endocrinology and Metabolism, 78: 308-315.
- 31. Gregersen H, Ritting S, Kraglund K & Phillips S (1988). The effect of glucagon and glucagon-(1-21)-peptide on antroduodenal motility in healthy volunteers. Scandinavian Journal of Gastroenterology, 152: 42-47.
- 32. MacLeod KM, Rodgers RL & McNeill JH (1981). Characterization of glucagon-induced changes in rate, contractility and cyclic AMP levels in isolated cardiac preparations of the rat and guinea pig. Journal of Pharmacology and Experimental Therapeutics, 217: 798-804.
- 33. Santamaria L, De Miguel E, Codesal J, Ramirez JR & Picazo J (1991). Identification of glucagon binding sites on smooth muscle tissue of dog intestine; quantification by means of ultrastructural autoradiography. Anatomischer Anzeiger, 172: 149-157.
- 34. Van Schravendijk CFH, Foriers A, Hooghe-Peters EL, Rogiers V, De Meyts P, Sodoyez JC & Pipeleers DG (1985). Pancreatic hormone receptors on islet cells. Endocrinology, 117: 841-848.
- 35. Kofod H, Unson C & Merrifield RB (1988). Potentiation of glucose-induced insulin release in islets by desHis1[Glu9]glucagon amide. International Journal of Peptide and Protein Research, 32: 436-440.
- 36. Abrahamsen N & Nishimura E (1995). Regulation of glucagon and glucagon-like peptide-1 receptor messenger ribonucleic acid expression in cultured rat pancreatic islets by glucose, cyclic adenosine 3',5'-monophosphate, and glucocorticoids. Endocrinology, 136: 1572-1578.
- 37. Bailly C, Imbert-Teboul M, Chabardés D, Hus-Citharel A, Montégut M, Clique A & Morel F (1980). The distal nephron of rat kidney: A target site for glucagon. Proceedings of the National Academy of Sciences, USA, 77: 3422-3424.
- 38. Delahouse M, Mercier O, Bichara M, Paillard M, Leviel F & Assan R (1988). Glucagon inhibits urinary acidification in the rat. American Journal of Physiology, 254: F762-F769.
- 39. Lok S, Kuijper JL, Jelinek LJ, Kramer JM, Whitmore TE, Sprecher CA, Mathewes S, Grant FJ, Biggs SH, Rosenberg GB, Sheppard PO, O'Hara PJ, Foster D & Kindsvogel W (1994). The human glucagon receptor encoding gene: structure, cDNA sequence and chromosomal localization. Gene, 140: 203-209.
- 40. Jelinek LJ, Lok S, Rosenberg GB, Smith RA, Grant FJ, Biggs S, Bensch PA, Kuijper JL, Sheppard PO, Sprecher CA, O'Hara PJ, Foster D, Walker KM, Chen LHJ, McKernan PA & Kindsvogel W (1993). Expression cloning and signaling properties of the rat glucagon receptor. Science, 259: 1614-1616.
- 41. Svoboda M, Ciccarelli E, Tastenoy M, Robberecht P & Christophe J (1993). A cDNA construct allowing the expression of rat hepatic glucagon receptors. Biochemical and Biophysical Research Communications, 192: 135-142.
- 42. Unson CG, Cypess AM, Kim HN, Goldsmith PK, Carruthers CJ, Merrifield RB & Sakmar TP (1995). Characterization of deletion and truncation mutants of the rat glucagon receptor. Seven transmembrane segments are necessary for receptor transport to the plasma membrane and glucagon binding. Journal of Biological Chemistry, 270: 27720-27727.
- 43. MacNeil DJ, Occi JL, Hey PJ, Strader CD & Graziano MP (1994). Cloning and expression of a human glucagon receptor. Biochemical and Biophysical Research Communications, 198: 328-334.
- 44. Wakelam MJ, Murphy GJ, Hruby VJ & Houslay MD (1986). Activation of two signal-transduction systems in hepatocytes by glucagon. Nature, 323: 68-71.
- 45. Iyengar R & Herberg JT (1984). Structural analysis of the hepatic glucagon receptor. Identification of a guanine nucleotide-sensitive hormone-binding region. Journal of Biological Chemistry, 259: 5222-5229.
- 46. Padrell E, Herberg JT, Monsatirsky B, Floyd G, Premont RT & Iyengar R (1987). The hepatic glucagon receptor: a comparative study of the regulatory and structural properties. Endocrinology, 120: 2316-2325.
- 47. Shimada M & Watanabe M (1993). The techniques and applications of whole-body radioautography. In: Nagata T (Editor), Radioautography in Medicine Shinshu University Press, Matsumoto, Japan, 33-40.
- 48. Kuhar MJ (1991). Perspectives on receptor autoradiography in human brain. In: Mendelsohn FAD & Paxinos G (Editors), Receptors in the Human Nervous System Academic Press, San Diego.
- 49. Curtis CG, Cross SAM, McCulloch RJ & Powell GM (1981). Whole-Body Autoradiography Academic Press, London.
- 50. Shimada M & Watanabe M (1995). Recent progress in whole-body radioautography. Cellular and Molecular Biology, 41: 39-48.
- 51. Rogers AW & Watanabe M (1988). The relative sensitivities of autoradiographic emulsions to negative chemography. Journal of Microscopy, 149: 193-197.
- 52. Watanabe M & Rogers AW (1987). Prevention of negative chemography in phenylenediamine stained autoradiographs of plastic semithin sections. Stain Technology, 62: 173-177.
- 53. Watanabe M, Koide T, Hayasaki H, Kanbara K, Jo N & Shimada M (1994). Negative chemography with mouse skeletal muscle. Bulletin of the Osaka Medical College, 40: 17-19.
- 54. Segu L, Rage P, Pinard R & Boulenguez P (1992). Computer-assisted quantitative receptor autoradiography. In: Conn PM (Editor), Computers and Computations in the Neurosciences Academic Press, San Diego, 129-143.
- 55. Watanabe M, Hirose Y & Shimada M (1991). A new technique for autoradiography of whole-body sections mounted on glass slides. Acta Histochemica et Cytochemica, 24: 167-171.
- 56. Shimono R, Watanabe M & Goto H (1982). A new method of preparation of whole-body sections for autoradiography and staining. Kaibogaku Zasshi, 57: 15-21.
- 57. Watanabe M, Kihara T, Shimada M & Kurimoto K (1978). Preparation and staining of whole-body sections. Cellular and Molecular Biology, 23: 311-315.
- 58. Watanabe M, Shimada M & Kurimoto K (1975). Staining method for whole-body autoradiography. Stain Technology, 50: 239-243.
- 59. Kuno N, Watanabe M, Akagi N, Sugimoto M & Kihara T (1987). A technique of autoradiography for tubular organs. Acta Histochemica et Cytochemica, 20: 547-555.
- 60. Rogers AW (1979). Techniques of Autoradiography 3rd edn. Elsevier North-Holland, Amsterdam.
- 61. Rogers AW (1979). Practical Autoradiography Review No. 20. Amersham International Limited, Amersham, UK.
- 62. Watanabe M, Hirose Y, Sugimoto M, Nakanishi M, Watanabe H & Shimada M (1992). The distribution of tissue insulin receptors in the mouse by whole-body autoradiography. Journal of Receptor Research, 12: 13-37.
- 63. Watanabe M, Kawamoto S, Nakatsuka Y, Koide T & Shimada M (1993). An application of the laser scanning microscope to microautoradiography. Acta Histochemica et Cytochemica, 26: 93-99.
- 64. Watanabe M, Koide T, Konishi M, Kanbara K & Shimada M (1995). Use of confocal laser scanning microscopy in radioautographic study. Cellular and Molecular Biology, 41: 131-136.
- 65. Watanabe M, Koide T, Nakatsuka Y & Shimada M (1993). Visualization of microradioautography by confocal laser scanning microscope. In: Nagata T (Editor), Radioautography in Medicine Shinshu University Press, Matsumoto, Japan, 215-219.
- 66. Watanabe M, Nishimura Y, Jo N, Takafuchi M, Kiyokane K & Shimada M (1996). Statistical analysis for receptor autoradiography. Acta Histochemica et Cytochemica, 29: 135-139.
- 67. Nishimura Y, Watanabe M, Jo N & Shimada M (1997). The difference of means of two indirect unpaired groups in receptor autoradiography. Journal of Biopharmaceutical Statistics, 7: 441-452.
- 68. Bergeron JJM, Levine G, Sikstrom R, O'Shaughnessy D, Kopriwa B, Nadler NJ & Posner BI (1977). Polypeptide hormone binding sites in vivo Initial localization of 125I-labeled insulin to hepatocyte plasma membrane as visualized by electron microscope radioautography. Proceedings of the National Academy of Sciences, USA, 74: 5051-5055.
- 69. Bergeron JJM, Sikstrom R, Hand AR & Posner BI (1979). Binding and uptake of 125I-insulin into rat liver hepatocytes and endothelium. An in vivo radioautographic study. Journal of Cell Biology, 80: 427-443.
- 70. Nakanishi M, Watanabe M & Shimada M (1995). Heterogeneous distribution of 125I-insulin binding sites in the liver of fed and fasted mice. Cellular and Molecular Biology, 41: 137-144.
- 71. Bergeron JJM, Rachubinski R, Searle N, Borts D, Sikstrom R & Posner BI (1980). Polypeptide hormone receptors in vivo: Demonstration of insulin binding to adrenal gland and gastrointestinal epithelium by quantitative radioautography. Journal of Histochemistry and Cytochemistry, 28: 824-835.
- 72. Shimada M & Watanabe M (1997). Whole-body and microradioautography of receptors. In: Morel G (Editor), Visualization of Receptors: Methods in Light and Electron Microscopy CRC Press, Paris, 97-124.
- 73. Whitcomb DC, O'Dorison TM, Cataland S, Shetzline MA & Nishikawara MT (1985). Identification of tissue insulin receptors: use of a unique in vivo radioreceptor assay. American Journal of Physiology, 249: E561-E567.
- 74. Bergeron JJM, Rachubinski R, Searle N, Sikstrom R, Borts D, Bastian P & Posner BI (1980). Radioautographic visualization of in vivo insulin binding to the exocrine pancreas. Endocrinology, 107: 1069-1080.
- 75. Sakamoto C, Williams JA, Roach E & Goldfine ID (1984). In vivo localization of insulin binding to cells of the rat pancreas. Proceedings of the Society for Experimental Biology and Medicine, 175: 497-502.
- 76. Dorn A, Bernstein HG, Hahn AJ, Zeigler M & Rummelfanger H (1981). Insulin immunohistochemistry of rodent CNS: apparent species differences but good correlation with radioimmunochemical data. Histochemistry, 71: 609-616.
- 77. Pansky B & Hatfield JS (1978). Cerebral localization of insulin by immunofluorescence. American Journal of Anatomy, 153: 459-467.
- 78. Baskin DG, Brewitt B, Davidson DA, Corp E, Paquette T, Figlewicz DP, Lewellen T, Graham MK, Woods SG & Dorsa DM (1986). Quantitative autoradiographic evidence for insulin receptors in the choroid plexus of the rat brain. Diabetes, 35: 246-249.
- 79. Baskin DG, Davidson DA, Corp E, Lewellen T & Graham M (1986). An inexpensive microcomputer digital imaging system for densitometry: quantitative autoradiography of insulin receptors with 125I and LKB Ultrofilm. Journal of Neuroscience Methods, 16: 119-129.
- 80. Hill JM, Lesniak MA, Pear CB & Roth J (1986). Autoradiographic localization of insulin receptors in rat brain: prominence in olfactory and limbic areas. Neuroscience, 17: 1127-1138.
- 81. Werther GA, Hogg A, Oldfield BJ, McKinley MJ, Figdor R, Allen M & Mendelsohn FAO (1987). Localization and characterization of insulin receptors in rat brain and pituitary gland using in vitro autoradiography and computerized densitometry. Endocrinology, 121: 1562-1570.
- 82. Marks JL, Porte Jr D, Stahl WL & Baskin DG (1990). Localization of insulin receptor mRNA in rat brain by in situ hybridization. Endocrinology, 127: 3234-3236.
- 83. Shimada M, Hirose Y, Nakanishi M, Watanabe H, Kawamoto S & Watanabe M (1992). Autoradiographic studies on insulin receptor of the mouse hippocampus. In: Wegmann RJ & Wegmann MA (Editors), Recent Advances in Cellular and Molecular Biology Vol. 4. Peeters Press, Leuven, Belgium, 271-278.
- 84. Martineau Doizé B, Warshawsky MH & Bergeron JJM (1986). In vivo demonstration by radioautography of binding sites for insulin in liver, kidney, and calcified tissues of the rat. Anatomical Record, 214: 130-140.
- 85. Sechi LA, De Carli S & Bartoli E (1994). In situ characterization of renal insulin receptors in the rat. Journal of Receptor Research, 14: 347-356.
- 86. Hirose Y, Watanabe M, Koide T & Shimada M (1994). Insulin-binding sites in the mouse deferent duct. Acta Histochemica, 96: 405-414.
- 87. Vinters HV & Berliner JA (1987). The blood vessel wall as an insulin target tissue. Diabete et Metabolisme, 13: 294-300.
- 88. Bar RS, Boes M & Sandra A (1988). Vascular transport of insulin to rat cardiac muscle. Central role of the capillary endothelium. Journal of Clinical Investigation, 81: 1225-1233.
- 89. Goldstein RH, Poliks CF, Pilch PF, Smith BD & Fine A (1989). Stimulation of collagen formation by insulin and insulin-like growth factor I in cultures of human lung fibroblasts. Endocrinology, 124: 964-970.
- 90. Loubassou S, Grizard G & Grizard J (1989). Insulin binding sites in solubilized membranes from rat testis. Effect of age. Andrologia, 21: 377-383.
- 91. Saucier J, Dube JY & Tremblay RR (1981). Specific insulin binding sites in rat testis: characterization and variation. Endocrinology, 109: 2220-2225.
- 92. Abele V, Pelletier G & Tremblay RR (1986). Radioautographic localization and regulation of the insulin receptors in rat testis. Journal of Receptor Research, 6: 461-473.
- 93. Carvalho CR, Brenelli SL, Silva AC, Nunes AL, Velloso LA & Saad MJ (1996). Effect of aging on insulin receptor, insulin receptor substrate-1, and phosphatidylinositol 3-kinase in liver and skeletal muscle. Endocrinology, 137: 151-159.
- 94. Munoz P, Rosemblatt M, Testar X, Palacin M, Thoidid G, Pilch PF & Zorzano A (1995). The T-tubule is a cell surface target for insulin-regulated recycling of membrane proteins in skeletal muscle. Biochemical Journal, 312: 393-400.
- 95. Jo N, Watanabe M, Kiyokane K & Shimada M (1997). In vivo microradioautographic study of insulin binding in the skin of normal and NIDDM mice: with special reference to acanthosis nigricans. Cellular and Molecular Biology, 43: 157-164.
- 96. Desoye G, Hartmann M, Blaschitz A, Dohr G, Hahn T, Kohnen G & Kaufmann P (1994). Insulin receptors in syncytiotrophoblast and fetal endothelium of human placenta. Immunohistochemical evidence for developmental changes in distribution pattern. Histochemistry, 101: 277-285.
- 97. Wolf HJ & Desoye G (1993). Immunohistochemical localization of glucose transporters and insulin receptors in human fetal membranes at term. Histochemistry, 100: 379-385.
- 98. Hyndmann AG (1995). Identification of a population of amacrine cells rich in insulin receptors. Brain Research, Developmental Brain Research, 75: 289-292.
- 99. Iwanij V (1995). Canine kidney glucagon receptor: evidence for a structurally-different, tissue-specific variant of the glucagon receptor. Molecular and Cellular Endocrinology, 115: 21-28.
- 100. Iwanij V & Vincent AC (1990). Characterization of the glucagon receptor and its functional domains using monoclonal antibodies. Journal of Biological Chemistry, 265: 21302-21308.
- 101. Hoosein NM & Gurd RS (1984). Identification of glucagon receptors in rat brain. Proceedings of the National Academy of Sciences, USA, 81: 4368-4372.
- 102. Montaron A, Moyse E & Barré H (1994). Radioautographic demonstration and localization of glucagon receptors in duck brain. Brain Research, 663: 121-130.
- 103. Koh WS, Lee M, Yang KH & Kaminski NE (1996). Expression of functional glucagon receptors on lymphoid cells. Life Sciences, 58: 741-751.
- 104. Frandsen EK & Bacchus RA (1987). Glucagon receptor binding, dissociation and degradation in rat liver plasma membranes studied by a microperfusion method. Biochimica et Biophysica Acta, 929: 74-80.
- 105. Berthould VM, Iwanij V, Garcia AM & Saez JC (1992). Connexins and glucagon receptors during development of rat hepatic acinus. American Journal of Physiology, 263: G650-G658.
- 106. Watanabe J, Kanamura S, Asada-Kubota M, Kanai K & Oka M (1984). Receptor-mediated endocytosis of glucagon in isolated mouse hepatocytes. Anatomical Record, 210: 557-567.
- 107. Watanabe J, Kanai K & Kanamura S (1988). Glucagon receptors in endothelial and Kupffer cells of mouse liver. Journal of Histochemistry and Cytochemistry, 36: 1081-1089.
- 108. Clark Jr CM, Beatty B & Allen DO (1973). Evidence for delayed development of the glucagon receptor of adenylate cyclase in the fetal and neonatal rat heart. Journal of Clinical Investigation, 52: 1018-1025.
- 109. Robberect P, Damien C, Moroder L, Coy DH, Wunsch E & Christophe J (1988). Receptor occupancy and adenylate cyclase activation in rat liver and heart membranes by 10 glucagon analogs modified in position 2, 3, 4, 25, 27 and/or 29. Regulatory Peptides, 21: 117-128.
- 110. Yount EA, Clark JF & Clark Jr CM (1976). Development of guanylylimidodiphosphate-dependent activation of adenylate cyclase by glucagon in the neonatal rat heart. Pediatric Research, 10: 851-854.
- 111. Desbuquois B & Authier F (1989). Glucagon receptors. Annales d' Endocrinologie, 50: 440-446.
- 112. Kawai K, Yokota C, Ohashi S, Watanabe Y & Yamashita K (1995). Evidence that glucagon stimulates insulin secretion through its own receptor in rats. Diabetologia, 38: 274-276.
- 113. Hansen LH, Abrahamsen N & Nishimura E (1995). Glucagon receptor mRNA distribution in rat tissues. Peptides, 16: 1163-1166.
- 114. Svoboda M, Tastenoy M, Vertongen P & Robberecht P (1994). Relative quantitative analysis of glucagon receptor mRNA in rat tissues. Molecular and Cellular Endocrinology, 105: 132-137.
- 115. Burcelin R, Li J & Charron MJ (1995). Cloning and sequence analysis of the murine glucagon receptor-encoding gene. Gene, 164: 305-310.
- 116. Christophe J (1994). Molecular cloning and functional expression of glucagon receptors in the liver, the heart and various other tissues. Bulletin et Memoires de l' Academie Royale de Medecine de Belgique, 149: 271-277.
- 117. Yoo-Warren H, Willse AG, Hancock N, Hull J, McCaleb M & Livingston JN (1994). Regulation of rat glucagon receptor expression. Biochemical and Biophysical Research Communications, 205: 347-353.
- 118. Camps M, Guma A, Vinals F, Testar X, Palacin M & Zorzano A (1992). Evidence for the lack of spare high-affinity insulin receptors in skeletal muscle. Biochemical Journal, 285: 993-999.
- 119. Joost HG (1995). Structural and functional heterogeneity of insulin receptors. Cell Signal, 7: 85-91.
Correspondence and Footnotes
Publication Dates
-
Publication in this collection
07 Oct 1998 -
Date of issue
Feb 1998
History
-
Accepted
22 July 1997 -
Received
11 July 1997