Abstract
Centromere function requires the proper coordination of several subfunctions, such as kinetochore assembly, sister chromatid cohesion, binding of kinetochore microtubules, orientation of sister kinetochores to opposite spindle poles, and their movement towards the spindle poles. Centromere structure appears to be organized in different, separable domains in order to accomplish these functions. Despite the conserved nature of centromere functions, the molecular genetic definition of the DNA sequences that form a centromere in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, in the fruit fly Drosophila melanogaster, and in humans has revealed little conservation at the level of centromere DNA sequences. Also at the protein level few centromere proteins are conserved in all of these four organisms and many are unique to the different organisms. The recent analysis of the centromere structure in the yeast S. pombe by electron microscopy and detailed immunofluorescence microscopy of Drosophila centromeres have brought to light striking similarities at the overall structural level between these centromeres and the human centromere. The structural organization of the centromere is generally multilayered with a heterochromatin domain and a central core/inner plate region, which harbors the outer plate structures of the kinetochore. It is becoming increasingly clear that the key factors for assembly and function of the centromere structure are the specialized histones and modified histones which are present in the centromeric heterochromatin and in the chromatin of the central core. Thus, despite the differences in the DNA sequences and the proteins that define a centromere, there is an overall structural similarity between centromeres in evolutionarily diverse eukaryotes.
Centromere; Kinetochore; Histone acetylation; Electron microscopy
Braz J Med Biol Res, May 2002, Volume 35(5) 499-507 (Review)
Centromere domain organization and histone modifications
P. Bjerling and K. Ekwall
Karolinska Institute, Sodertorn University College, Huddinge, Sweden
Text
References
Correspondence and Footnotes Correspondence and Footnotes Correspondence and Footnotes
Abstract
Centromere function requires the proper coordination of several subfunctions, such as kinetochore assembly, sister chromatid cohesion, binding of kinetochore microtubules, orientation of sister kinetochores to opposite spindle poles, and their movement towards the spindle poles. Centromere structure appears to be organized in different, separable domains in order to accomplish these functions. Despite the conserved nature of centromere functions, the molecular genetic definition of the DNA sequences that form a centromere in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, in the fruit fly Drosophila melanogaster, and in humans has revealed little conservation at the level of centromere DNA sequences. Also at the protein level few centromere proteins are conserved in all of these four organisms and many are unique to the different organisms. The recent analysis of the centromere structure in the yeast S. pombe by electron microscopy and detailed immunofluorescence microscopy of Drosophila centromeres have brought to light striking similarities at the overall structural level between these centromeres and the human centromere. The structural organization of the centromere is generally multilayered with a heterochromatin domain and a central core/inner plate region, which harbors the outer plate structures of the kinetochore. It is becoming increasingly clear that the key factors for assembly and function of the centromere structure are the specialized histones and modified histones which are present in the centromeric heterochromatin and in the chromatin of the central core. Thus, despite the differences in the DNA sequences and the proteins that define a centromere, there is an overall structural similarity between centromeres in evolutionarily diverse eukaryotes.
Key words: Centromere, Kinetochore, Histone acetylation, Electron microscopy
Introduction
The centromere is the region of the chromosome that is responsible for its segregation at mitosis and meiosis. Centromeres are therefore essential for genetic stability and there are several examples where defects in centromere function are associated with aneuploidy (aberrant chromosome number) causing birth defects or with cancer. Centromeres have been molecularly defined in several genetic model organisms and several centromere proteins have been characterized (for reviews, see Refs. 1,2). In this review we present a survey of centromere DNA sequences and centromere proteins from yeasts, Drosophila and humans. We focus on the comparison of centromere domain structure in the light of recent cytological characterization of Drosophila and Schizosaccharomyces pombe centromeres (3,4). We also address the exciting mechanistic insights regarding the involvement of histone modifications in centromere assembly and function.
Centromeric DNA sequences
A survey of centromere DNA sequences from yeast, Drosophila and humans is presented in Table 1. In human cells the centromeric DNA consists of AT-rich a-satellite repeats (5). However, this DNA sequence is not absolutely required for centromere assembly and function since so-called `neocentromeres' can sometimes be formed in regions of the chromosome which lack a-satellite DNA (6,7). In Drosophila, the centromere has been assigned on a Dp1187 minichromosome to a 420-kb region rich in repetitive DNA sequences (8). Also in Drosophila neocentromeres can in some cases be formed on other centromere sequences (9). Thus, both in human and Drosophila cells there is plasticity with respect to the DNA sequences that can form a centromere. In fission yeast (S. pombe) the centromere DNA encompasses 40- to 100-kb regions of the chromosome and has a symmetric organization. The minimal DNA sequence requirement for an S. pombe centromere is a central core sequence (CC/cnt) flanked by a portion of repeated arrays that consist of inner (imr/B) and outer (otr/K + L) sequences (10-12). In budding yeast, centromeres are relatively small and 120 bp of DNA is sufficient for a fully functional centromere (13). Thus, at least in humans and Drosophila the exact DNA sequences are not important for the formation of a functional centromere and in general there is no conservation between species with regard to the DNA sequences that can form a centromere.
Centromere proteins
A survey of centromere proteins from two yeasts, Drosophila and humans is presented in Table 1. Several centromere proteins have been defined in humans through the binding of autoantibodies from human patients with scleroderma. These immunoglobulins react with several distinct centromere proteins (CENP) (14,15). In Drosophila many centromere proteins have been defined: MEI-S332, POLO, PROD, ROD, ZW10, BUB1, BUB3, CENP-meta, HP1 and CID (DmCENP-A) (16-24). These proteins are part of different subdomains of the Drosophila centromere: the pericentric heterochromatin, the inner plate and the outer kinetochore (3). In S. pombe several centromere proteins have also been identified: Swi6, Chp1, Cnp1, Mis6, Mis12, Ndc80, Nuf2, Spc24, Alp14/Dis1 and Mtc1 (25-34). Chromatin immunoprecipitation crosslinking experiments have demonstrated that these proteins are also part of different centromeric subdomains. For example, Cnp1 (S. pombe CENP-A) and Mis6 proteins both bind to the central core region but not to the flanking regions (27,29,30). Conversely, the chromodomain proteins Swi6 and Chp1 bind to the flanking repeats but not to the central core region (35). This indicates that there are at least two distinct structural and functional domains in S. pombe centromeres. In budding yeast, centromere proteins have been defined and extensively studied. Recently, budding yeast kinetochore proteins were re-defined using cytological live analysis and dynamic criteria (36). A total of 11 proteins including the S. cerevisiae counterparts of Nuf2, Ndc80, Spc24 and Spc25 were defined as kinetochore proteins in this study. The kinetochore proteins are likely to be anchored to the centromere via a DNA-binding protein complex CBF3 (37) and Cse4, which is the budding yeast CENP-A homologue, forming a specialized nucleosome which in turn forms the centromeric chromatin (38). Notably the pericentric heterochromatin is lacking in budding yeast centromeres and the corresponding heterochromatin proteins are missing. The S. cerevisiae centromeres are thus smaller in size, indicating that the heterochromatin component may have been lost during the evolution of these `trimmed down' kinetochores. To summarize, several centromere proteins have now been defined in the various organisms and although many factors appear to be organism specific it is possible to discern some degree of conservation (underlined proteins in Table 1). Interestingly, the variant histone CENP-A at the inner plate/central core seems to be completely conserved and parts of the Ndc80 protein complex (although hitherto not described in Drosophila) seem to be a conserved component of most eukaryotic kinetochores.
Centromere domain structure
Recent studies have revealed that the positions of some centromere proteins are conserved within the multilayered kinetochore structures from S. pombe to humans (Figure 1). In S. pombe the centromere proteins Mis6 and Cnp1 (SpCENP-A) associate exclusively with central core DNA, while the Swi6 protein binds to the surrounding repeats, suggesting that distinct protein interaction domains exist within the S. pombe centromeres (35). Electron microscopy and immunofluorescence light microscopy of the precisely positioned centromeres in interphase cells have revealed that the central core and flanking regions indeed occupy cytologically distinct positions within a heterochromatic domain. In addition, an `anchor' structure containing the Ndc80 putative kinetochore protein was found between this heterochromatic domain and the spindle pole body (4). As seen by electron microscopy, the human metaphase centromere is multilayered, containing several substructures: a fibrous corona, an outer and an inner plate, as well as the space between them (39). Beneath the inner plates is the heterochromatic region. Each of these substructures appears to comprise a distinct protein composition. The fibrous corona contains CENP-E, dynein and dynactin, the outer plate contains CENP-F, the inner plate contains CENP-C and CENP-A, and the underlying heterochromatin contains CENP-B, INCENP, HP1 and SUV39H1 (40-46). Live analysis of cells expressing CENP-A and CENP-B green fluorescent protein fusion proteins indicates that, although the human kinetochore unfolds and refolds during interphase, the human pre-kinetochore structure remains ordered in interphase. CENP-A localization is limited to the edge of a larger CENP-B heterochromatin domain even before the typical double dot structure appears in G2 (47,48). At metaphase, CENP-A is a component of the inner plate in the human centromere (44). The human homologue of Ndc80, HEC, is localized to the outer part of HeLa cell centromeres (31). There are also human homologues of additional components of the budding yeast kinetochore, i.e., XMAP215, CLIP170 and BimC kinesins but their localization in the human centromere has not been determined (36).
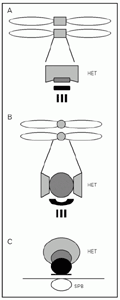 Drosophila (B), and the centromere cluster near the SPB at interphase in Schizosaccharomyces pombe (C). The heterochromatic (HET) domains (light gray), the inner kinetochore/inner plate/central core structures (dark gray) and the outer plate/outer kinetochore/anchor structures (black) are indicated. The vertical lines from the outer plate/kinetochore in A and B represent kinetochore microtubules. SPB = spindle pole body.
Drosophila (B), and the centromere cluster near the SPB at interphase in Schizosaccharomyces pombe (C). The heterochromatic (HET) domains (light gray), the inner kinetochore/inner plate/central core structures (dark gray) and the outer plate/outer kinetochore/anchor structures (black) are indicated. The vertical lines from the outer plate/kinetochore in A and B represent kinetochore microtubules. SPB = spindle pole body.
[View larger version of this image (20 K GIF file)]
Histone modifications
Several different covalent modifications of the histone proteins have been reported to affect centromeric chromatin. These are acetylation, methylation and phosphorylation. Acetylation of histones is carried out by histone acetyl transferases at several conserved lysine residues of the N-terminal tails of histone H3 and H4. In general, histones in heterochromatin are relatively underacetylated when compared to those present in euchromatin (49,50). The opposite reaction, the deacetylation of histone tails, is performed by histone deacetylases (51). For S. pombe, Drosophila and human cells it has been shown that the centromeres are underacetylated (50,52). Treatment of S. pombe and human cells with the histone deacetylase inhibitor trichostatin A increases acetylation in centromeric heterochromatin and disrupts centromere function (50,53). The underacetylated state of the flanking repeats is necessary for binding of Swi6 to S. pombe centromeres and for binding of HP1 to human pericentric heterochromatin (50,53). Swi6 has been shown to associate with centromeric heterochromatin in fission yeast, and this association has been shown to require Clr4 (25,54). Recently, Clr4 belonging to a conserved chromo- and SET-domain family of proteins was shown to modify the N-terminal tail of histone H3 by methylation (55,56). Indeed H3 methylation was shown to be required for Swi6 binding to centromeric otr/K chromatin which is methylated at lysine 9 of histone H3 (57,58). It has been shown that deacetylation of histone H3 by histone deacetylases is required to allow for the subsequent methylation of this residue by the Clr4 histone H3 methyltransferase in a stepwise mechanism (57). The mouse SUV39H1 is a homologue of Clr4 with the same enzymatic function. It has been shown to be required for chromosome segregation and interacts with M31 (mouse HP1 homologue) in pericentric heterochromatin (46). SUV39H1 specifically accumulates in the centromere during prometaphase but dissociates from centromeric positions in anaphase (59). Su(var)3-9 in Drosophila was originally defined as a gene required for position effect variegation, i.e., silencing of the centromeric marker gene but it remains to be tested whether the protein itself is associated with centromeres. Thus, underacetylation of histones H3 and H4 and methylation of lysine 9 of histone H3 are required for heterochromatin formation and/or proper chromosome segregation in several different organisms. What is then the exact function of centromeric heterochromatin?
It is known from S. pombe that trichostatin A-induced acetylated centromeres display lagging chromosomes in anaphase cells. This phenotype was recently shown to be the consequence of a defective centromeric cohesion function since the cohesin Rad21 is dependent on Swi6 for its association with this region (60). Thus, although this remains to be demonstrated directly, it is conceivable that underacetylation of centromeric outer domain at least in S. pombe is a prerequisite for the centromeric cohesion function. Yet additional modifications are known to occur in centromeric histones. Phosphorylation occurs both of "normal histone H3" and of the histone H3 variant, CENP-A, localized to the central core of the centromere (61). In human cells this phosphorylation has been shown to occur in three phases of the cell cycle. Pericentric regions first accumulate H3 phosphorylation, followed by general phosphorylation of all chromatin and lastly by phosphorylation of CENP-A in the centromere (61). Interestingly, the Aurora B kinase colocalizes with INCENP at meta-phase chromosomes and is responsible for phosphorylation of histone H3 as well as being required for metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation (62,63). Notably the budding yeast homologue of Aurora kinase, Ipl1, is present in the kinetochore of this organism (36). The exact targets of the Ipl1 kinase at the budding yeast kinetochores are currently not known but Ipl1 is required for segregation of sister chromatids at anaphase (64).
Kinetochore assembly
The domain organization of the centromere described in Drosophila (3) is similar to the one described in S. pombe and humans. These findings now allow us to address the function of the different centromere domains in the genetic model organisms Drosophila and S. pombe. To this end, the study of Blower and Karpen indicated that CENP-A (CID) has a very central role in several centromere functions. Injection of antibodies against CID and/or RNAi inhibition of CID expression resulted in a multitude of centromeric defects including prophase arrest, indicative of an assembly defect, and lagging chromosomes which is indicative of cohesion defect. Inhibition of CID led to delocalization of several centromere proteins (POLO, ROD, BUB1, CENP-meta) indicating that the incorporation of all these factors is normally dependent on CID. In budding yeast Cse4 is necessary for proper chromosome segregation. In a cse4 conditional mutant, the core centromere chromatin structure is disrupted at the restrictive temperature (38,65). Similarly, in fission yeast a mutation in the mis6+ (mis-segregation) gene causes a failure of the Cnp1 (SpCENP-A) protein to bind the central core chromatin leading to massive aneuploidy (29,30). Although it remains to be investigated whether other kinetochore components are lost from centromeres in mis6-1 cells, the strong defects indicate that CENP-A may be required for kinetochore assembly also in S. pombe. What targets CENP-A to centromeres? Replication timing might be important to form a functional centromere and has been suggested as an important determinant for CENP-A targeting (66). In Drosophila it has been shown that the centromeres replicate in the early S-phase while surrounding heterochromatin replicates later (67). This replication pattern might be crucial since a human neocentromere 10q25.3 causes the surrounding DNA to replicate later in the S-phase as compared to a wild-type chromosome 10 (68). However, other investigations show that replication timing does not matter for the incorporation of CENP-A and formation of a functional centromere (69). In fission yeast otr/K repeats in the centromeres replicate early (70) and CENP-A is expressed early in the S-phase (30). However, there is no evidence for an early replication requirement for CENP-A incorporation into S. pombe centromeres.
Conclusions and perspectives
Centromere DNA sequences were recently defined molecularly in humans and can be compared with those of genetic model organisms, i.e., two evolutionarily distant yeasts (S. pombe and S. cerevisiae) and Drosophila. Perhaps surprisingly, given the crucial function of centromeres in maintaining genomic stability, there is no obvious conservation of the DNA sequences which form centromeres in the different eukaryotes. Even the centromere proteins are in many cases species-specific but comparative approaches have revealed some key components such as the histone H3 variant CENP-A which is required for centromere function in all systems. Despite the differences in DNA sequences and protein composition of centromeres, the structural organization at the electron microscope level reveals a conserved multilayered domain organization between S. pombe, Drosophila and humans (Figure 1). These centromeres are based on a heterochromatic part of the chromosome that contains a special chromatin structure with underacetylated histones. It is conceivable that this structural similarity is conserved to accommodate the common subfunctions of a centromere: kinetochore assembly, sister chromatid cohesion, binding of kinetochore microtubules, orientation of sister kinetochores to opposite spindle poles, and their movement towards the spindle poles.
Address for correspondence: K. Ekwall, Karolinska Institute, Sodertorn University College, Box 4101, S-141 04 Huddinge, Sweden. Fax: +46-8-585-88-510. E-mail: karl.ekwall@cbt.ki.se
Presented at the XXX Annual Meeting of the Brazilian Society of Biochemistry and Molecular Biology, Caxambu, MG, Brazil, May 19-22, 2001. Research supported by Cancerfonden (grant #4284-B01-03XAB) and the Swedish Medical Research Council (grant #K00-31X-12562- 03A) to K. Ekwall. Received January 14, 2002. Accepted February 19, 2002.
- 1. Pluta AF, Mackay AM, Ainsztein AM, Goldberg IG & Earnshaw WC (1995). The centromere: hub of chromosomal activities. Science, 270: 1591-1594.
- 2. Pidoux AL & Allshire RC (2000). Centromeres: getting a grip of chromosomes. Current Opinion in Cell Biology, 12: 308-319.
- 3. Blower MD & Karpen GH (2001). The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nature Cell Biology, 3: 730-739.
- 4. Kniola B, O'Toole E, McIntosh JR, Mellone B, Allshire R, Mengarelli S, Hultenby K & Ekwall K (2001). The domain structure of centromeres is conserved from fission yeast to humans. Molecular Biology of the Cell, 12: 2767-2775.
- 5. Schueler MG, Higgins AW, Rudd MK, Gustashaw K & Willard HF (2001). Genomic and genetic definition of a functional human centromere. Science, 294: 109-115.
- 6. Cancilla MR, Tainton KM, Barry AE, Larionov V, Kouprina N, Resnick MA, Sart DD & Choo KH (1998). Direct cloning of human 10q25 neocentromere DNA using transformation-associated recombination (TAR) in yeast. Genomics, 47: 399-404.
- 7. Depinet TW, Zackowski JL, Earnshaw WC, Kaffe S, Sekhon GS, Stallard R, Sullivan BA, Vance GH, Van Dyke DL, Willard HF, Zinn AB & Schwartz S (1997). Characterization of neo-centromeres in marker chromosomes lacking detectable alpha-satellite DNA. Human Molecular Genetics, 6: 1195-1204.
- 8. Sun X, Wahlstrom J & Karpen G (1997). Molecular structure of a functional Drosophila centromere. Cell, 91: 1007-1019.
- 9. Williams BC, Murphy TD, Goldberg ML & Karpen GH (1998). Neocentromere activity of structurally acentric mini-chromosomes in Drosophila Nature Genetics, 18: 30-37.
- 10. Clarke L, Amstutz H, Fishel B & Carbon J (1986). Analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe Proceedings of the National Academy of Sciences, USA, 83: 8253-8257.
- 11. Clarke L & Baum MP (1990). Functional analysis of a centromere from fission yeast: a role for centromere-specific repeated DNA sequences. Molecular and Cellular Biology, 10: 1863-1872.
- 12. Chikashige Y, Kinoshita N, Nakaseko Y, Matsumoto T, Murakami S, Niwa O & Yanagida M (1989). Composite motifs and repeat symmetry in S. pombe centromeres: direct analysis by integration of NotI restriction sites. Cell, 57: 739-751.
- 13. Cottarel G, Shero JH, Hieter P & Hegemann JH (1989). A 125-base-pair CEN6 DNA fragment is sufficient for complete meiotic and mitotic centromere functions in Saccharomyces cerevisiae Molecular and Cellular Biology, 9: 3342-3349.
- 14. Earnshaw WC & Migeon BR (1985). Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma, 92: 290-296.
- 15. Earnshaw WC & Rothfield N (1985). Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma, 91: 313-321.
- 16. Kerrebrock AW, Moore DP, Wu JS & Orr-Weaver TL (1995). Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell, 83: 247-256.
- 17. Logarinho E & Sunkel CE (1998). The Drosophila POLO kinase localises to multiple compartments of the mitotic apparatus and is required for the phosphorylation of MPM2 reactive epitopes. Journal of Cell Science, 111: 2897-2909.
- 18. Torok T, Harvie PD, Buratovich M & Bryant PJ (1997). The product of proliferation disrupter is concentrated at centromeres and required for mitotic chromosome condensation and cell proliferation in Drosophila Genes and Development, 11: 213-225.
- 19. Scaerou F, Aguilera I, Saunders R, Kane N, Blottiere L & Karess R (1999). The rough deal protein is a new kinetochore component required for accurate chromosome segregation in Drosophila Journal of Cell Science, 112: 3757-3768.
- 20. Williams BC, Gatti M & Goldberg ML (1996). Bipolar spindle attachments affect redistributions of ZW10, a Drosophila centromere/kinetochore component required for accurate chromosome segregation. Journal of Cell Biology, 134: 1127-1140.
- 21. Basu J, Logarinho E, Herrmann S, Bousbaa H, Li Z, Chan GK, Yen TJ, Sunkel CE & Goldberg ML (1998). Localization of the Drosophila checkpoint control protein Bub3 to the kinetochore requires Bub1 but not Zw10 or Rod. Chromosoma, 107: 376-385.
- 22. Yucel JK, Marszalek JD, McIntosh JR, Goldstein LS, Cleveland DW & Philp AV (2000). CENP-meta, an essential kinetochore kinesin required for the maintenance of metaphase chromosome alignment in Drosophila Journal of Cell Biology, 150: 1-11.
- 23. Saunders WS, Chue C, Goebl M, Craig C, Clark RF, Powers JA, Eissenberg JC, Elgin SC, Rothfield NF & Earnshaw WC (1993). Molecular cloning of a human homologue of Drosophila heterochromatin protein HP1 using anti-centromere autoantibodies with anti-chromo specificity. Journal of Cell Science, 104: 573-582.
- 24. Henikoff S, Ahmad K, Platero JS & van Steensel B (2000). Heterochromatic deposition of centromeric histone H3-like proteins. Proceedings of the National Academy of Sciences, USA, 97: 716-721.
- 25. Ekwall K, Javerzat JP, Lorentz A, Schmidt H, Cranston G & Allshire R (1995). The chromodomain protein Swi6: a key component at fission yeast centromeres. Science, 269: 1429-1431.
- 26. Doe CL, Wang G, Chow C, Fricker MD, Singh PB & Mellor EJ (1998). The fission yeast chromo domain encoding gene chp1(+) is required for chromosome segregation and shows a genetic interaction with alpha-tubulin. Nucleic Acids Research, 26: 4222-4229.
- 27. Goshima G, Saitoh S & Yanagida M (1999). Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes and Development, 13: 1664-1677.
- 28. Bernard P, Hardwick K & Javerzat JP (1998). Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. Journal of Cell Biology, 143: 1775-1787.
- 29. Saitoh S, Takahashi K & Yanagida M (1997). Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell, 90: 131-143.
- 30. Takahashi K, Chen ES & Yanagida M (2000). Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science, 288: 2215-2219.
- 31. Wigge PA & Kilmartin JV (2001). The Ndc80 complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. Journal of Cell Biology, 152: 349-360.
- 32. Nabetani A, Koujin T, Tsutsumi C, Haraguchi T & Hiraoka Y (2001). A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma, 110: 322-334.
- 33. Garcia MA, Vardy L, Koonrugsa N & Toda T (2001). Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO Journal, 20: 3389-3401.
- 34. Nakaseko Y, Goshima G, Morishita J & Yanagida M (2001). M phase-specific kinetochore proteins in fission yeast: microtubule-associating Dis1 and Mtc1 display rapid separation and segregation during anaphase. Current Biology, 11: 537-549.
- 35. Partridge JF, Borgstrom B & Allshire RC (2000). Distinct protein interaction domains and protein spreading in a complex centromere. Genes and Development, 14: 783-791.
- 36. He X, Rines DR, Espelin CW & Sorger PK (2001). Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell, 106: 195-206.
- 37. Lechner J & Carbon J (1991). A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell, 64: 717-725.
- 38. Meluh PB, Yang P, Glowczewski L, Koshland D & Smith MM (1998). Cse4p is a component of the core centromere of Saccharomyces cerevisiae Cell, 94: 607-613.
- 39. Pluta AF, Cooke CA & Earnshaw WC (1990). Structure of the human centromere at metaphase. Trends in Biochemical Sciences, 15: 181-185.
- 40. Cooke CA, Schaar B, Yen TJ & Earnshaw WC (1997). Localization of CENP E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma, 106: 446-455.
- 41. Yao X, Anderson KL & Cleveland DW (1997). The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. Journal of Cell Biology, 139: 435-447.
- 42. Saitoh H, Tomkiel J, Cooke CA, Ratrie 3rd H, Maurer M, Rothfield NF & Earnshaw WC (1992). CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell, 70: 115-125.
- 43. Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, Poirier G & Earnshaw WC (1997). Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Current Biology, 7: 901-904.
- 44. Vafa O & Sullivan KF (1997). Chromatin containing CENP A and alpha satellite DNA is a major component of the inner kinetochore plate. Current Biology, 7: 897-900.
- 45. Cooke CA, Bernat RL & Earnshaw WC (1990). CENP-B: a major human centromere protein located beneath the kinetochore. Journal of Cell Biology, 110: 1475-1488.
- 46. Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, Reuter G & Jenuwein T (1999). Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO Journal, 18: 1923-1938.
- 47. Pudenko AS, Kudryavtsev IS, Zatsepina OV & Chentsov Yu S (1997). Spatial association of prekinetochores and chromocentres in the interphase nuclei of mouse cultured fibroblasts. Membrane and Cell Biology, 11: 449-461.
- 48. Sugimoto K, Fukuda R & Himeno M (2000). Centromere/kinetochore localization of human centromere protein A (CENP-A) exogenously expressed as a fusion to green fluorescent protein. Cell Structure and Function, 25: 253-261.
- 49. Jeppesen P & Turner BM (1993). The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell, 74: 281-289.
- 50. Ekwall K, Olsson T, Turner BM, Cranston G & Allshire RC (1997). Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell, 91: 1021-1032.
- 51. Taunton J, Hassig CA & Schreiber SL (1996). A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science, 272: 408-411.
- 52. Turner BM, Birley AJ & Lavender J (1992). Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell, 69: 375-384.
- 53. Taddei A, Maison C, Roche D & Almouzni G (2001). Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nature Cell Biology, 3: 114-120.
- 54. Ekwall K, Nimmo ER, Javerzat JP, Borgstrom B, Egel R, Cranston G & Allshire R (1996). Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. Journal of Cell Science, 109: 2637-2648.
- 55. Ivanova AV, Bonaduce MJ, Ivanov SV & Klar AJ (1998). The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nature Genetics, 19: 192-195.
- 56. Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD & Jenuwein T (2000). Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature, 406: 593-599.
- 57. Nakayama J, Rice JC, Strahl BD, Allis CD & Grewal SI (2001). Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science, 292: 110-113.
- 58. Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC & Kouzarides T (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410: 120-124.
- 59. Aagaard L, Schmid M, Warburton P & Jenuwein T (2000). Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. Journal of Cell Science, 113: 817-829.
- 60. Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP & Allshire RC (2001). Requirement of heterochromatin for cohesion at centromeres. Science, 294: 2539-2542.
- 61. Zeitlin SG, Barber CM, Allis CD & Sullivan K (2001). Differential regulation of CENP-A and histone H3 phosphorylation in G2/M. Journal of Cell Science, 114: 653-661.
- 62. Adams RR, Eckley DM, Vagnarelli P, Wheatley SP, Gerloff DL, Mackay AM, Svingen PA, Kaufmann SH & Earnshaw WC (2001). Human INCENP colocalizes with the Aurora-B/AIRK2 kinase on chromosomes and is overexpressed in tumour cells. Chromosoma, 110: 65-74.
- 63. Adams RR, Maiato H, Earnshaw WC & Carmena M (2001). Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. Journal of Cell Biology, 153: 865-880.
- 64. Biggins S, Bhalla N, Chang A, Smith DL & Murray AW (2001). Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae Genetics, 159: 453-470.
- 65. Stoler S, Keith KC, Curnick KE & Fitzgerald-Hayes M (1995). A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes and Development, 9: 573-586.
- 66. Shelby RD, Vafa O & Sullivan KF (1997). Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. Journal of Cell Biology, 136: 501-513.
- 67. Ahmad K & Henikoff S (2001). Centromeres are specialized replication domains in heterochromatin. Journal of Cell Biology, 153: 101-110.
- 68. Lo AW, Craig JM, Saffery R, Kalitsis P, Irvine DV, Earle E, Magliano DJ & Choo KH (2001). A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO Journal, 20: 2087-2096.
- 69. Sullivan B & Karpen G (2001). Centromere identity in Drosophila is not determined in vivo by replication timing. Journal of Cell Biology, 154: 683-690.
- 70. Kim SM & Huberman JA (2001). Regulation of replication timing in fission yeast. EMBO Journal, 20: 6115-6126.
Correspondence and Footnotes
Publication Dates
-
Publication in this collection
06 May 2002 -
Date of issue
May 2002
History
-
Accepted
19 Feb 2002 -
Received
14 Jan 2002


