Abstract
Saccharomyces cerevisiae mutants deficient in superoxide dismutase genes (sod1delta, sod2delta and the double mutant) were subjected to H2O2 stress in the stationary phase. The highest sensitivity was observed in the sod2delta mutant, while the sod1deltasod2delta double mutant was not sensitive. Sod mutants had lower catalase activity (44%) than wild-type cells, independent of H2O2 stress. Untreated cells of sod1deltasod2delta double mutants showed increased glutathione peroxidase activity (126%), while sod1delta had lower activity (77%) than the wild type. Glutathione levels in sod1delta were increased (200-260%) after exposure to various H2O2 concentrations. In addition, the highest malondialdehyde levels could be observed without H2O2 treatment in sod1delta (167%) and sod2delta (225%) mutants. In contrast, the level of malondialdehyde in the sod1deltasod2delta double mutant was indistinguishable from that of the wild type. These results suggest that resistance to H2O2 by sod1deltasod2delta cells depends on the induction of glutathione peroxidase and is independent of catalase, and that glutathione is a primary antioxidant in the defense against H2O2 in stationary phase sod1delta mutants.
Catalase; Superoxide dismutase; Glutathione; Hydrogen peroxide; Saccharomyces cerevisiae; Reactive oxygen species
Braz J Med Biol Res, February 2004, Volume 37(2) 159-165
Glutathione peroxidase induction protects Saccharomyces cerevisiae sod1 D sod2 D double mutants against oxidative damage
V. Manfredini1, R. Roehrs2, M.C.R. Peralba3, J.A.P. Henriques1,2, J. Saffi2, A.L.L.P. Ramos1 and M.S. Benfato1
1Laboratório de Estresse Oxidativo, Departamento de Biofísica, 2Centro de Biotecnologia, and 3Departamento de Química Inorgânica, Instituto de Química, Universidade Federal do Rio Grande do Sul, Porto Alegre, RS, Brasil
References
Correspondence and Footnotes Correspondence and Footnotes Correspondence and Footnotes
Abstract
Saccharomycescerevisiae mutants deficient in superoxide dismutase genes (sod1D, sod2D and the double mutant) were subjected to H2O2 stress in the stationary phase. The highest sensitivity was observed in the sod2D mutant, while the sod1Dsod2D double mutant was not sensitive. Sod mutants had lower catalase activity (44%) than wild-type cells, independent of H2O2 stress. Untreated cells of sod1Dsod2D double mutants showed increased glutathione peroxidase activity (126%), while sod1D had lower activity (77%) than the wild type. Glutathione levels in sod1D were increased (200-260%) after exposure to various H2O2 concentrations. In addition, the highest malondialdehyde levels could be observed without H2O2 treatment in sod1D (167%) and sod2D (225%) mutants. In contrast, the level of malondialdehyde in the sod1Dsod2D double mutant was indistinguishable from that of the wild type. These results suggest that resistance to H2O2 by sod1Dsod2D cells depends on the induction of glutathione peroxidase and is independent of catalase, and that glutathione is a primary antioxidant in the defense against H2O2 in stationary phase sod1D mutants.
Key words: Catalase, Superoxide dismutase, Glutathione, Hydrogen peroxide, Saccharomyces cerevisiae, Reactive oxygen species
Introduction
Oxygen metabolism may lead to the production of reactive oxygen species (ROS), i.e., superoxide and hydroxyl radicals and hydrogen peroxide (H2O2), by sequential one-electron reductions. ROS damages all cellular components, including protein, DNA and lipids. A primary source of superoxide is the electron transport chain in the inner membrane of mitochondria, where about 2% of the oxygen consumed during respiration is incompletely reduced to ROS. To counteract the oxidative stress resulting from ROS, cells possess a range of nonenzymatic and enzymatic defense systems, including glutathione (GSH), thioredoxin, superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPx) (1).
A first line of defense includes the SOD enzymes that catalyze the disproportionate cleavage of superoxide to H2O2 and water. H2O2 is enzymatically catabolized in aerobic organisms by catalase and several peroxidases. Saccharomyces cerevisiae, like most other eukaryotes, contains CuZnSOD (the product of the SOD1 gene) in the cytosol, nucleus, and lysosomes and MnSOD (the product of the SOD2 gene) in the mitochondrial matrix.
MnSOD is believed to be the major means of protection against mitochondrial superoxide (2). The sources of ROS relevant to CuZnSOD are less clear because of the location of this enzyme within the cytosol. Neither MnSOD nor CuZnSOD is strictly essential; however, the loss of CuZnSOD has dramatic phenotypic consequences in yeast. Yeast strains lacking CuZnSOD (sod1D) show several defects during aerobic growth. These include reduced growth rates in glycerol and ethanol, auxotrophy for lysine and methionine or cysteine, a higher rate of spontaneous mutation, more rapid loss of viability in the stationary phase, increased iron uptake, elevated levels of "free iron", and sensitivity to redox-cycling drugs such as paraquat or menadione (3-8). sod2D mutants are oxygen-sensitive and, when required to utilize oxygen, grow poorly and are particularly sensitive to paraquat (6). sod1Dsod2D double mutants are more severely affected, exhibiting essentially all the characteristics of the single mutant phenotypes.
The full growth cycle of a yeast culture begins with the exponential phase of growth (log phase) and progresses through the diauxic shift to the true stationary phase. In the log phase, the cells use glucose to produce energy via glycolysis. The diauxic shift occurs when fermentable nutrients become limited and energy metabolism shifts to respiration, the growth rate slows, and cells utilize ethanol and other two- and three-carbon compounds for energy production. In the total absence of any nutrients, the cells enter the true stationary phase; no cell division occurs, the metabolic rate slows and cells can survive for weeks to months.
Stationary phase yeast resemble the majority of cells of multicellular organisms in two important aspects: 1) their main source of energy is mitochondrial respiration, and 2) the cells have exited the cell cycle and entered the G0 phase. With increasing time in the stationary phase, damage accumulates. This damage cannot be excluded by cell synthesis and division, which is not occurring, and thus must be prevented or repaired. For this reason the yeast S. cerevisiae has been extensively exploited as a model for advancing our understanding of cellular defenses against ROS.
In the present study we investigated the role of nonenzymatic and enzymatic defense systems after H2O2 treatment in sod mutants. We found that sod1D and sod1Dsod2D double mutants show very little or no sensitivity to H2O2 in the stationary phase. The double mutant displays increased GPx activity and reduced malondialdehyde (MDA) levels compared to wild-type cells, whereas CAT is decreased. The sod1D mutant has the highest total GSH content after H2O2. Thus, the resistance to H2O2 in sod1Dsod2D cells is dependent on the induction of GPx, whereas CAT does not appear to be required. We suggest here that GSH is a primary antioxidant in the defense against H2O2 in sod1D mutants.
Material and Methods
Strains of Saccharomycescerevisiae
The wild-type S. cerevisiae strain EG103 and the isogenic mutant strains sod1D, sod2D and sod1Dsod2D were kindly provided by Dr. E. Gralla (University of California, Los Angeles, CA, USA). Disruption of the SOD1 and SOD2 genes was performed as described previously (4,9). MG5312 was a gift from Dr. M. Brendel (J.W. Goethe-University, Frankfurt, Germany). The relevant genotypes of the strains are listed in Table 1.
Glutathione peroxidase induction protects Saccharomyces cerevisiae sod1Dsod2D double mutants against oxidative damage. V. Manfredini, R. Roehrs, M.C.R. Peralba, J.A.P. Henriques, J. Saffi, A.L.L.P. Ramos and M.S. Benfato. Brazilian Journal of Medical and Biological Research, 37 (2): 159, 2004.
Media and growth conditions
Yeast strains were grown at 30ºC in YPD liquid medium containing 2% glucose, 1% yeast extract, 2% bacto-peptone, or selective medium supplemented with the appropriate nutrients (SD medium with 2% glucose, 0.67% yeast nitrogen base without amino acids, plus nutrients). The flask volume/medium ratio was 2:1 (microaerophilic conditions) and flasks were shaken at 200 rpm. For solid medium 2% agar was added (10). The cell lines lacking CuZnSOD were continually monitored for suppressor activity (4,6).
Hydrogen peroxide treatment
Yeast cells were grown to the stationary phase (1-2 x 108 cells/ml) in YPD medium at 30ºC. Cells were harvested and washed in sterile saline (0.9% NaCl) and the cell pellets were resuspended in saline and treated with increasing concentrations of H2O2 (0.5-10 mM) at 30ºC for 1 h.
For dose-response curves, aliquots of cells were diluted in saline and plated in triplicate onto YPD to obtain viable counts after 3-5 days of growth at 30ºC.
Enzyme activities
Crude extracts were prepared by glass bead lysis as follows: cells were suspended in lysis buffer (50 mM Tris, 150 mM NaCl, 50 mM EDTA, pH 7.2) with an equal volume of acid-washed 425-600 µm glass beads and phenylmethylsulfonyl fluoride, vortexed for 10-15 cycles (30 s each), followed by 30 s of cooling. The mixture was then microcentrifuged for 2 min to remove the cellular debris and glass beads (6).
CAT activity was determined spectrophotometrically by monitoring the disappearance of H2O2 at 240 nm (11). GPx activity was determined by monitoring the NADPH consumption rate at 340 nm (12). Protein concentration was determined by the Bradford assay (13).
Assay of total glutathione
Total GSH was monitored by a recently developed microbiological method that uses a GSH auxotrophic yeast strain as a sensor for the presence of GSH, GSSG and g-Glu-Cys (14). The indicator strain, S. cerevisiae MG5312 (Table 1), homozygous for the mutant allele gsh1DURA3, was grown to a cell density of 2 x 108 cells/ml, washed twice with and then suspended in potassium phosphate buffer (25 mM KH2PO4, 50 mM Na2HPO4, pH 7.0) to a concentration of 2 x 107 cells/ml. One hundred microliters of this suspension was added to 3.5 ml top-agar (0.4% agar in potassium phosphate buffer) at a temperature of 48ºC and poured immediately onto synthetic medium lacking GSH. After the top-agar had solidified, 20 µl of the samples to be tested was applied to a sterile paper disc 10 mm in diameter on the agar surface. Plates were incubated at 30ºC for 3 days. Growth of MG5312 depends on the presence either GSH or GSSG. Growth zones on agar plates seeded with MG5312 permit the quantitative determination as little as 0.1 µg GSH (for details see Ref. 14).
Preparationofyeastsamples. After H2O2 treatment yeast cells were harvested, washed twice and suspended in potassium phosphate buffer to a final concentration of 4%. Cell suspensions were heat-treated for 15 min at 85ºC and cell debris was removed by centrifugation at 11,700 g.
Malondialdehyde determination
MDA was measured by HPLC by the method described by Esterbauer and Cheeseman (15). Briefly, an aqueous sample containing MDA, pH 6.5-8.0, was separated by HPLC using an amino-phase column with acetonitrile, 30 mM Tris buffer, pH 7.4 (1:9 v/v). The effluent was monitored at 267 nm (15).
Statistical analysis
Results are reported as means ± SD and were analyzed by the Student t-test. Values of P < 0.05 were considered to be statistically significant.
Results
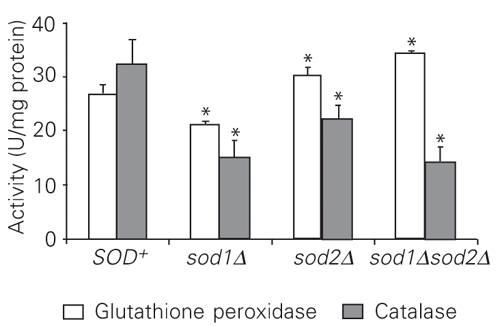
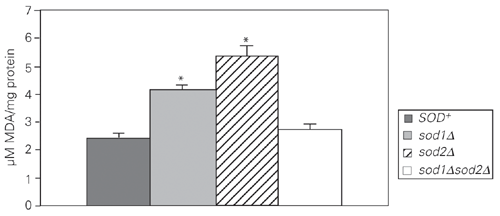
Wild-type and sod null mutant cultures in the stationary phase of growth under microaerophilic conditions were treated with increasing levels of H2O2 for 1 h at 30ºC under non-growth conditions. The results (Figure 1) showed that the sod2D mutant was more sensitive to H2O2 than its isogenic parent strain, while the sensitivity of sod1D was similar to that of the wild type. Surprisingly, the sensitivity of the sod1Dsod2D double mutant was indistinguishable from that of the wild type. In order to determine the possible mechanisms that may contribute to H2O2 resistance in the sod1Dsod2D and sod1D mutants, antioxidant enzyme activities were assayed (Figures 2-4). Decreased CAT activity was observed either with or without H2O2 treatment in simple and double sod mutants (Figures 2 and 3). On the other hand, GPx activities were significantly increased in sod2D and sod1Dsod2D mutants without treatment, while sod1D showed lower GPx activity than the wild type (Figure 2). After H2O2 treatment, GPx levels were significantly increased when exposed to 0.25-0.5 mM H2O2 and strongly reduced at 2.5 mM H2O2 in the wild-type and mutant strains (Figure 4). At the highest doses of H2O2 (5-10 mM), GPx levels were the same as in untreated cells for sod2D, while wild-type and sod1Dsod2D levels increased over control cell levels and sod1D maintained lower GPx activities. The total GSH levels in these mutants were increased at 0.5 mM H2O2 and were reduced 6- to 7-fold at 1 mM H2O2 in the wild type, while the strain lacking both SODs showed a reduction in GSH levels both with and without treatment (Figure 5). The single sod1D mutant showed increased GSH at 0.5 to 5 mM H2O2. Little increase in the GSH level was caused by H2O2 treatment in the single sod2D mutant. The lipid peroxidation index was determined on the basis of MDA levels (Figure 6). The highest MDA levels were observed without H2O2 treatment in simple sod mutants. On the other hand, the MDA level was indistinguishable from that of the wild type in the sod1Dsod2D double mutant.
Sensitivity of sod null mutants exposed to H2O2. The strains used were: SOD+, sod1D, sod2D and sod1Dsod2D. Data are reported as the mean ± SD of three independent experiments.
Activity of catalase and glutathione peroxidase in untreated wild-type and sod mutant cells. Data are reported as the mean ± SD of three independent experiments. *P < 0.05 compared to SOD+ (Student t-test).
Activity of catalase in stationary wild-type and sod mutant cells exposed to H2O2.The strains used were: SOD+, sod1D, sod2D and sod1Dsod2D. Data are reported as the mean ± SD of three independent experiments. aP < 0.05 compared to SOD+-untreated cells; bP < 0.05 compared to sod1D-untreated cells; cP < 0.05 compared to sod2D-untreated cells; dP < 0.05 compared to sod1Dsod2D-untreated cells (Student t-test).
Activity of glutathione peroxidase in wild-type and sod mutant cells exposed to H2O2. The strains used were: SOD+, sod1D, sod2D and sod1Dsod2D. Data are reported as the mean ± SD of three independent experiments. aP < 0.05 compared to SOD+-untreated cells; bP < 0.05 compared to sod1D-untreated cells; cP < 0.05 compared to sod2D-untreated cells; dP < 0.05 compared to sod1Dsod2D-untreated cells (Student t-test).
Total glutathione (GSH) content of wild-type and sod mutant cells exposed to H2O2. The total GSH was monitored by a microbiological method that uses a GSH auxotrophic yeast strain as a sensor for the presence of GSH, GSSG and g-Glu-Cys. Data are reported as the mean ± SD of two independent experiments. aP < 0.05 compared to SOD+-untreated cells; bP < 0.05 compared to sod1D-untreated cells; cP < 0.05 compared to sod2D-untreated cells; dP < 0.05 compared to sod1Dsod2D-untreated cells (Student t-test).
Malondialdehyde (MDA) determination in untreated wild-type and sod mutant cells. Data are reported as the mean ± SD of four independent experiments. *P < 0.05 compared to SOD+ (Student t-test).
Discussion
Metabolically active cells consume oxygen intensively; under limited aeration, this leads to low oxygen concentration within the cells. In stationary cultures, metabolism is considerably reduced and the partial pressure of oxygen can increase as a result of decreased metabolism. Under these conditions of high oxidative stress, one-electron reactions of autoxidation-prone cellular components may produce superoxide and other ROS. Mitochondrial respiration has been suggested to be the major source of ROS under these conditions, even or especially under conditions of low aeration.
Oxidative stress may be a factor limiting the survival of microorganisms in long-term stationary culture. The increases in antioxidant content and in the levels of antioxidant enzymes, including SOD, in yeast entering the stationary phase may, therefore, constitute an adaptive response to the enhanced oxidative damage (16). The resistance to H2O2 in sod1Dsod2D cells is dependent on the induction of GPx rather than CAT, the latter apparently being unnecessary. In the absence only of MnSOD (sod2D mutant), GPx was induced, but this did not prevent cell mortality (70% at 10 mM H2O2; Figure 1). However, the cells lacking CuZnSOD (sod1D mutant) were resistant to H2O2 (Figure 1). These results cannot be explained by GPx or CAT induction, because both GPx and CAT activities were lower in the sod1D mutant (Figures 2-4).
We also investigated the levels of total GSH in these mutants and in the wild type (Figure 5). The sod1D mutant showed increased GSH after H2O2 treatment. These results indicate that GSH is a primary antioxidant in the defense against H2O2 in sod1D cells. Mixed disulfides of GSSG with proteins accumulate in tissues subjected to oxidative stress, in both the mitochondria and the cytosol (17). GSSG can inhibit protein synthesis in animal and plant cells (18,19). These actions of GSSG may explain why cells keep intracellular GSSG levels very low under normal conditions, and why cells export GSSG when they are under oxidative stress (20-23). The sod1Dsod2D double mutant showed reduced total GSH levels with or without H2O2 treatment (Figure 5), while a slight increase in GSH levels was observed in the sod2D mutant after H2O2 treatment. We therefore suggest that GSSG and/or mixed disulfides are exported by these cells. It is known that GSSG levels are similar in exponential phase cells and their media during growth conditions and that the extracellular GSH level is elevated following treatment with H2O2 concentrations greater than 0.5 mM (24). We obtained similar results in the stationary phase (Figure 5).
The highest levels of antioxidant defenses, including GPx or total GSH, were observed in sod mutants. CAT levels, however, were lower and the enzyme was not induced after H2O2 treatment. SOD enzymes catalyze the disproportionate cleavage of superoxide, producing oxygen and H2O2; so sod mutants should contain less H2O2. H2O2 is enzymatically detoxified within the cells by CAT and GPx; however, GPx also detoxifies other peroxides. The acatalasemic mutant cells in the stationary phase were much more sensitive to H2O2 stress than wild-type cells. In addition, the ability of acatalasemic cells to show adaptation to H2O2 treatment has been shown to be distinctly inferior to that of the wild type (25). These results suggest that CAT is not essential for yeast cells under normal conditions, but plays an important role in the acquisition of tolerance to oxidative stress in the adaptive response. The sod mutants did not induce CAT in spite of stress in their intracellular environment (Figure 3). If superoxide is not adequately removed it could inhibit CAT (26). However, sod mutants induced GPx (Figure 4).
These results suggest that sod mutants produce other type(s) of intracellular peroxides. It has been shown recently that the GPx genes of S. cerevisiae encode phospholipid hydroperoxide GPx and that these enzymes protect yeast against phospholipid hydroperoxides as well as nonphospholipid peroxides during oxidative stress (27,28). Therefore, we can suggest that single sod mutants have increased lipid peroxidation as well as protein damage (7,8). Our results showed that lack of CuZnSOD or MnSOD causes a significant increase, 71 and 124%, respectively, in the levels of MDA, an indicator of lipid peroxidation (Figure 6). However, cells lacking both CuZnSOD and MnSOD enzymes did not show elevated MDA levels. These findings are in agreement with the GPx induction and resistance to H2O2 observed in sod1Dsod2D.
Acknowledgments
The authors wish to thank Dr. C. Gaylard for a critical English revision, and Dr. C.R. Carlini for generously providing equipment.
Address for correspondence: M.S. Benfato, Departamento de Biofísica, Instituto de Biociências, UFRGS, Av. Bento Gonçalves, 9500, Prédio 43422, 91501-970 Porto Alegre, RS, Brasil. Fax: +55-51-3316-7003. E-mail: mbenfato@adufrgs.ufrgs.br
Research supported by FAPERGS, CNPq and GENOTOX-CBIOT-UFRGS. Received April 25, 2003. Accepted August 28, 2003.
- 1. Halliwell B & Gutteridge JMC (1999). Free Radicals in Biology and Medicine 3rd edn. Oxford University Press, New York.
- 2. Gralla EB & Kosman DJ (1992). Molecular genetics of superoxide dismutases in yeasts and related fungi. Advances in Genetics, 30: 251-319.
- 3. Farr SB, D'Ari R & Touati D (1986). Oxygen-dependent mutagenesis in Escherichia coli lacking superoxide-dismutase. Proceedings of the National Academy of Sciences, USA, 83: 8268-8272.
- 4. Gralla EB & Valentine JS (1991). Null mutants of Saccharomyces cerevisiae Cu,Zn superoxide dismutase: characterization and spontaneous mutation rates. Journal of Bacteriology, 173: 5918-5920.
- 5. Bilinski T, Krawiec Z, Liczmanski A & Litwinska J (1985). Is hydroxyl radical generated by the Fenton reaction in vivo? Biochemical and Biophysical Research Communications, 130: 533-539.
- 6. Longo VD, Gralla EB & Valentine JS (1996). Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Journal of Biological Chemistry, 271: 12275-12280.
- 7. De Freitas JM, Liba A, Meneghini R, Valentinie JS & Gralla EB (2000). Yeast lacking superoxide dismutase show altered iron homeostasis. Role of oxidative stress in iron metabolism. Journal of Biological Chemistry, 275: 11645-11649.
- 8. Srinivasan C, Liba A, Imlay JA, Valentine JS & Gralla EB (2000). Yeast lacking superoxide dismutase(s) show elevated levels of "free iron" by whole cell electron paramagnetic resonance. Journal of Biological Chemistry, 275: 29187-29192.
- 9. Liu XF, Elashvili I, Gralla EB, Valentine JS, Lapinskas P & Culotta VC (1992). Yeast lacking superoxide-dismutase - isolation of genetic suppressors. Journal of Biological Chemistry, 267: 18298-18302.
- 10. Shermann F, Fink GR & Hicks JB (1986). Methods in Yeast Genetics Cold Spring Harbor, New York.
- 11. Taniguchi N & Gutteridge JMC (2000). Experimental Protocols for Reactive Oxygen and Nitrogen Species 1st edn. Oxford University Press, New York.
- 12. Pinto RE & Bartley W (1969). Effect of age and sex on GSHreductase and GSHperoxidase activities and on aerobic glutathione oxidation in rat liver homogenates. Biochemical Journal, 112: 109-115.
- 13. Bradford M (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72: 248-254.
- 14. Schmidt M, Grey M & Brendel M (1996). A microbiological assay for the quantitative determination of glutathione. BioTechniques, 21: 881-886.
- 15. Esterbauer H & Cheeseman KH (1990). Determination of aldehydic lipid peroxidation products: malonaldehyde and 4-hydroxynonenal. Methods in Enzymology, 186: 407-421.
- 16. Jakubowski W, Bilinski T & Bartosz G (2000). Oxidative stress during aging of stationary cultures of the yeast Saccharomyces cerevisiae Free Radical Biology and Medicine, 28: 659-664.
- 17. Ravichandran V, Seres T, Moriguchi T, Thomas JA & Johnston RB (1994). S-Thiolation of glyceraldehyde-3-phosphate dehydrogenase induced by the phagocytosis-associated respiratory burst in blood monocytes. Journal of Biological Chemistry, 269: 25010-25015.
- 18. Schuppekoistinen I, Gerdes R, Moldeus P & Cotgreave IA (1994). Studies on the reversibility of protein S-thiolation in human endothelial cells. Archives of Biochemistry and Biophysics, 315: 226-234.
- 19. Dhindsa RS (1987). Glutathione status and protein synthesis during drought and subsequent rehydration in Tortula ruralis Plant Physiology, 83: 816-919.
- 20. Hirrlinger J, Schulz JB & Dringen R (2002). Effects of dopamine on the glutathione metabolism of cultured astroglial cells. Implications for Parkinson's disease. Journal of Neurochemistry, 82: 458-467.
- 21. Wernerman J, Luo JL & Hammarqvist F (1999). Glutathione status in critically-ill patients; possibility of modulation by antioxidants. Proceedings of the Nutrition Society, 58: 677-680.
- 22. Spooren AAMG & Evelo CTA (1997). Hydroxylamine treatment increases glutathione-protein and protein-protein binding in human erythrocytes. Blood Cells, Molecules, and Diseases, 17: 323-336.
- 23. Lapshina EA & Bartosz G (1995). What determines the antioxidant potential of yeast cells. Biochemistry and Molecular Biology International, 37: 949-957.
- 24. Grant CM, Perrone G & Dawes IW (1998). Glutathione and catalase provide overlapping defenses for protection against hydrogen peroxide in the yeast Saccharomyces cerevisiae. Biochemical and Biophysical Research Comunications , 253: 893-898.
- 25. Izawa S, Inoue Y & Kimura A (1996). Importance of catalase in the adaptive response to hydrogen peroxide: analysis of acatalasaemic Saccharomyces cerevisiae. Biochemical Journal, 320: 61-67.
- 26. Lardinois OM (1995). Reactions of bovine liver catalase with superoxide radicals and hydrogen-peroxide. Free Radical Research, 22: 251-274.
- 27. Inoue Y, Matsuda T, Sugiyama K, Izawa S & Kimura A (1999). Genetic analysis of glutathione peroxidase in oxidative stress response of Saccharomyces cerevisiae. Journal of Biological Chemistry, 274: 27002-27009.
- 28. Avery AM & Avery SM (2001). Saccharomyces cerevisiae expresses three phospholipid hydroperoxide glutathione peroxidases. Journal of Biological Chemistry, 276: 33730-33735.
Correspondence and Footnotes
Publication Dates
-
Publication in this collection
30 Jan 2004 -
Date of issue
Feb 2004
History
-
Accepted
28 Aug 2003 -
Received
25 Apr 2003