Abstract
We report here the construction of a vector derived from pET3-His and pRSET plasmids for the expression and purification of recombinant proteins in Escherichia coli based on T7 phage RNA polymerase. The resulting pAE plasmid combined the advantages of both vectors: small size (pRSET), expression of a short 6XHis tag at N-terminus (pET3-His) and a high copy number of plasmid (pRSET). The small size of the vector (2.8 kb) and the high copy number/cell (200-250 copies) facilitate the subcloning and sequencing procedures when compared to the pET system (pET3-His, 4.6 kb and 40-50 copies) and also result in high level expression of recombinant proteins (20 mg purified protein/liter of culture). In addition, the vector pAE enables the expression of a fusion protein with a minimal amino-terminal hexa-histidine affinity tag (a tag of 9 amino acids using XhoI restriction enzyme for the 5'cloning site) as in the case of pET3-His plasmid and in contrast to proteins expressed by pRSET plasmids (a tag of 36 amino acids using BamHI restriction enzyme for the 5'cloning site). Thus, although proteins expressed by pRSET plasmids also have a hexa-histidine tag, the fusion peptide is much longer and may represent a problem for some recombinant proteins.
Escherichia coli; Expression vector; Immobilized metal affinity chromatography; Protein purification
Braz J Med Biol Res, August 2004, Volume 37(8) 1103-1109
A high-copy T7 Escherichia coli expression vector for the production of recombinant proteins with a minimal N-terminal His-tagged fusion peptide
C.R.R. Ramos1,2, P.A.E. Abreu1,2, A.L.T.O. Nascimento1,2 and P.L. Ho1,2,3
1Centro de Biotecnologia, Instituto Butantan, São Paulo, SP, Brasil
2Instituto de Química, and 3Instituto de Biociências, Universidade de São Paulo, São Paulo, SP, Brasil
 References
References
 Acknowledgments
Correspondence and Footnotes
Acknowledgments
Acknowledgments
Correspondence and Footnotes
Acknowledgments
Abstract
We report here the construction of a vector derived from pET3-His and pRSET plasmids for the expression and purification of recombinant proteins in Escherichia coli based on T7 phage RNA polymerase. The resulting pAE plasmid combined the advantages of both vectors: small size (pRSET), expression of a short 6XHis tag at N-terminus (pET3-His) and a high copy number of plasmid (pRSET). The small size of the vector (2.8 kb) and the high copy number/cell (200-250 copies) facilitate the subcloning and sequencing procedures when compared to the pET system (pET3-His, 4.6 kb and 40-50 copies) and also result in high level expression of recombinant proteins (20 mg purified protein/liter of culture). In addition, the vector pAE enables the expression of a fusion protein with a minimal amino-terminal hexa-histidine affinity tag (a tag of 9 amino acids using XhoI restriction enzyme for the 5'cloning site) as in the case of pET3-His plasmid and in contrast to proteins expressed by pRSET plasmids (a tag of 36 amino acids using BamHI restriction enzyme for the 5'cloning site). Thus, although proteins expressed by pRSET plasmids also have a hexa-histidine tag, the fusion peptide is much longer and may represent a problem for some recombinant proteins.
Key words: Escherichia coli, Expression vector, Immobilized metal affinity chromatography, Protein purification
Introduction
One of the most useful systems for expression of recombinant proteins in Escherichia coli is the pET vector series (Novagen, Madison, WI, USA), which is based on the T7 phage RNA polymerase promoter (1) and uses the pBR322 origin of DNA replication. The expression of the recombinant protein using these plasmids is tightly regulated and, when induced, produces high levels of transcripts and recombinant proteins (2). Several pET vectors have an affinity tag composed of six consecutive histidine residues (6XHis-tag) that permit the purification of the fusion protein on metal charged columns (3). In contrast, the pRSETA expression system (Invitrogen, Carlsbad, CA, USA) is a high-copy number plasmid based on pUC19 origin of DNA replication that may confer a higher gene dose effect (4). In this system, the proteins are produced with fusion of one leader peptide that encodes the first 12 amino acids of the T7 gene 10 major capsid protein (T7-epitope tag), and the affinity 6XHis-tag. An enterokinase cleavage site is located between the leader peptide and the cloning site and therefore the epitope and affinity tag can be excised off the recombinant protein if desired.
In many cases the single 6XHis-tag does not affect the structure and function of the recombinant protein. Moreover, the 6XHis-tag may be able to act as an epitope of protein detection using monoclonal antibodies or enzymenitrilotriacetic acidnickel conjugates.
In the present report we describe the construction of a vector for the production of heterologous proteins in E. coli that combines the characteristics of both pET vectors and pRSET expression systems. The resulting vector, named pAE, is a high copy number plasmid that allows the production of recombinant proteins with a minimal 6XHis-tag at the N-terminus.
Material and Methods
Bacterial strains, plasmids and growth conditions
The E. coli strain DH5a (Life Technologies, Inc., Rockville, MD, USA) was used for all routine cloning experiments whereas the E. coli strain BL21 (DE3) (Novagen) was used for recombinant protein expression. The pRSET vector series was purchased from Invitrogen, and the pET3-His (5) plasmid was kindly provided by the Cloning Vector Collection (National Institute of Genetics, Shizuota-ken, Japan). Luria Bertani medium (LB) was used for culture with a supplement of 100 µg ampicillin (Sigma, St. Louis, MO, USA) per ml.
Cloning techniques
All DNA manipulations were carried out as described in Ref. 6. The DNA fragment coding for the tetanus toxin fragment C (Fc) was amplified by the polymerase chain reaction (PCR) from pET32a-Fc (7) using the forward primer containing XhoI and BamHI sites and the reverse primer containing the HindIII site. PCR was carried out with 50 µl of reaction mixture containing 10 ng template DNA, 0.2 mM each dNTP, 20 pmol each primer, 1X PCR buffer, and 0.5 U Taq DNA polymerase (Life Technologies). The PCR assay was performed in a GeneAmp PCR system 9600 (Perkin Elmer, Foster City, CA, USA) for 30 cycles under the following conditions: 94ºC for 45 s, 56ºC for 25 s, and 72ºC for 3 min, followed by a final extension at 72ºC for 15 min. The amplified product was purified by agarose gel electrophoresis using a commercial extraction system (In Concert Gel Extraction System, Life Technologies). The recovered DNA was then cloned into the plasmid pET3-His at XhoI and HindIII sites, resulting in the pET3-His-Fc plasmid. Next, the DNA insert containing the 6XHis-tag and the Fc coding sequence was isolated from pET3-His-Fc by double digestion with NdeI and HindIII and further subcloned into the pRSETA vector at the same restriction sites. The resulting plasmid was named pRSET-His-Fc. The entire multiple cloning site and part of the pRSETC plasmid backbone were re-inserted at BamHI and ScaI sites by replacement of the Fc coding sequence plus the corresponding fragment of the pRSETA backbone, resulting in the pAE vector. All constructions were confirmed by sequencing in the ABI 377 automatic sequencer (PE Applied Biosystem, Foster City, CA, USA) using the ABI Prism Big Dye Terminator kit and T7 promoter primer.
Expression and purification of recombinant fragment C
The E. coli BL21 (DE3) cells transformed with pAE-Fc and pET3-His-Fc were grown at 37ºC in LB medium containing ampicillin (100 mg/l). When absorbance at 600 nmreached 0.6, isopropyl-1-thio-ß-D-galactoside (IPTG) was added at a final concentration of 1 mM and the cells were harvested by centrifugation 3 h later. Aliquots of total cell extracts were collected every 30 min and analyzed by 10% SDS-PAGE. For the purification of the recombinant protein, the bacterial pellet was resuspended in 20 mM Tris-HCl, pH 8.0, and lysed with a French Press. The inclusion bodies were collected by centrifugation. The solubilization buffer (20 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 6 M guanidine hydrochloride, 1% Triton X-100, and 2 mM ß-mercaptoethanol) was added to the pellet, incubated for 3 h at room temperature and clarified by centrifugation, and the supernatant was collected. Purification was performed by batch using Fast Flow chelating Sepharose (Pharmacia, Uppsala, Sweden) charged with nickel sulfate. The clarified supernatant was adsorbed to the charged resin and the resin was washed with 20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 6 M urea, 2 mM ß-mercaptoethanol, and 60 mM imidazole. The recombinant protein was then eluted with 1 M imidazole in the same buffer and characterized by 10% SDS-PAGE.
Results and Discussion
The pET vector series is derived from pBR322 (8). The plasmid yield from a typical miniprep is at least five times less when compared to pRSET vector series, which is derived from pUC19 (Figure 1) (9). As a result, manipulations of pET vectors often require multiple steps, like ethanol precipitation or speed vac concentration and growth of a larger amount of E. coli culture, in order to obtain the necessary amount and concentration of plasmid for vector digestion for subcloning and/or for direct plasmid sequencing. In contrast, the pRSET series yields a larger amount of plasmid, permitting direct digestion for subcloning and sequencing from a single 1.5-ml miniprep.
The pET3-His plasmid (5) is a pET-derived vector that permits the rapid purification of the recombinant protein through a minimal 6XHis-tag fused to the expressed protein at the N-terminus. In order to combine the advantages of the pRSET vectors with those of the pET3-His vector, we constructed the pAE plasmid. First, we cloned the tetanus toxin Fc into the pET3-His vector (Figure 2). The 1.35-kb DNA fragment coding for the Fc was amplified by PCR using the forward primer containing the XhoI and BamHI sites and the reverse primer with a HindIII site. The resulting DNA fragment was purified and cloned into the plasmid pET3-His at XhoI and HindIII sites, resulting in the pET3-His-Fc plasmid. Next, the DNA insert containing the 6XHis-tag and the Fc coding sequence was isolated from pET3-His-Fc by double digestion with NdeI and HindIII and further subcloned into the pRSETA vector at the same restriction sites. The resulting plasmid was named pAE-Fc. The entire multiple cloning site and part of the pRSETA plasmid backbone was re-inserted at BamHI and ScaI sites by replacement of the Fc coding sequence plus the corresponding piece of the pRSETA backbone, yielding the pAE vector (Figure 2). The pAE plasmid may combine the advantage of a minimal 6XHis-tag just following the ATG start codon from the pET3-His plasmid with the multiple cloning sites and the replicative origin of pRSET plasmid series (Figure 3).
To test if the resulting pAE plasmid would produce plasmid yields similar to those of the pRSET plasmid series, cultures of E. coli transformed with pRSETA, pET3-His or pAE were grown until absorbance at 600 nm reached 1.7. The plasmids from each culture were isolated using commercial kits (In Concert Rapid Plasmid Miniprep System, Life Technologies) and 2.5 µl of the plasmid DNA's were digested with BamHI and submitted to agarose gel electrophoresis. The DNA plasmids were visualized after staining with ethidium bromide (Figure 1) and quantified by densitometry using standard plasmid DNA (pGEM3zf(+), Promega, Madison, WI, USA) of known concentration. The same plasmid miniprep from each culture was also quantified by absorbance at 260 nm (Table 1). Both pAE and pRSETA plasmids gave similar yields, about 200-250 plasmid copies per cell, as estimated by absorbance at 260 nm and by fluorescence in an agarose gel (Figure 1 and Table 1). pET3-His, as expected, yielded five times less plasmid when compared to pAE and pRSETA. The pAE plasmid is smaller (2.8 kb), which would allow the cloning of larger DNA inserts than the pET3-His vector. The control of plasmid copy number resides in a region of the origin of DNA replication. The replication of ColE1 type plasmids is regulated by the interaction of RNA I and Rop (repressor of primer) protein with the primer for replication and RNA II (10) that is synthesized from a promoter located 555 bp upstream of the origin of DNA replication (11). The maturation of RNA II is controlled by another untranslated RNA molecule (RNA I), which is transcribed from the opposite strand of the same region of DNA coding for RNA II. In addition to the effect of RNA I on primer formation, the ColE1 copy number is modulated by a second plasmid-encoded element, the product of the rop gene, which consists of a 63-amino acid polypeptide (12). The pET vector series is based on the pBR322 plasmid that replicates through the pMB1 replicon whereas pRSETA is a pUC18/19 derived from a pBR322 replicon through mutations in RNA I and rop genes. This prevents the interaction of RNA I and Rop protein with RNA II, resulting in a higher plasmid copy number per cell. In theory, as a consequence, the higher copy number of an expression plasmid containing a pUC18/19 origin of DNA replication would result in a greater amount of expression of the recombinant protein due to a dosage effect, as was reported for the bovine growth hormone in E. coli (4). This could be another advantage of the pAE vector over the pET3-His plasmid in addition to the larger plasmid yield (Figure 2 and Table 1).
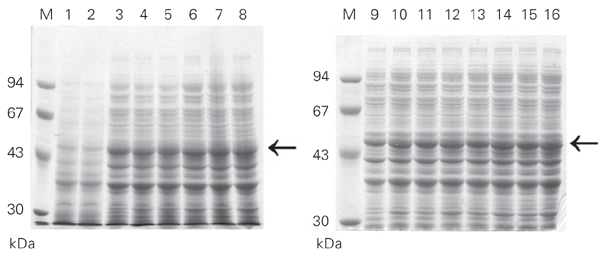
To test this possibility, we expressed the Fc using the pAE and the pET3-His vectors. Both vectors are able to express an identical recombinant Fc in terms of amino acid sequence and 6XHis-tag fusion. Transformed E. coli BL21 (DE3) cells with pAE-Fc or pET3-His-Fc were incubated at 37ºC until absorbance at 600 nm reached 0.6 and the cultures were induced with 1 mM IPTG as described. The cells were collected with time, lysed with SDS-PAGE sample buffer and analyzed by SDS-PAGE (Figure 4). Both pAE-Fc and pET3-His-Fc transformed cells expressed the same amount of recombinant Fc with time. In this case, the greater copy number/cell of the pAE-Fc plasmid over pET3-His-Fc (data not shown) did not result in a larger amount of expressed recombinant protein. The tetanus toxin Fc was purified from both pAE-Fc and pET3-His-Fc transformed cells using Ni2+-charged resin as described in Material and Methods. The Fc yields were similar from both cultures, around 20 mg/l of culture (Figure 5).
The advantage of the Histag over the GST or FLAGtag is that the fusion protein can be purified under denaturing conditions since the interaction between consecutive His residues and the affinity column, nickel chelating column, is stable in 6 M guanidine HCl or 8 M urea which are strong protein denaturants. The purified protein can then be renatured by removing the denaturant slowly. Since many foreign proteins produced in E. coli are insoluble, the Histag is more widely applicable than the GST or FLAGtag (13). The pAE vector described here allows the expression of heterologous proteins with a minimal His-tag placed just after the methionine start codon, like pET3-His and in contrast to pRSETA. While pET3-His and pAE permit the expression of a polypeptide with a minimal number of 9 amino acids (including the hexa-histidine tag) fused at the N-terminus (for instance, using XhoI as the cloning site), the minimal fusion peptide expressed by pRSETA without the use of enterokinase is composed of 36 amino acids (using BamHI as the cloning site; Figure 3). Although the use of enterokinase would remove the hexa-histidine tag, yielding a fusion peptide of 5 amino acids, this is not a predictable feature, depending on the exposure of the enterokinase cleavage site in the recombinant protein expressed by the pRSETA plasmid. Its use also includes additional steps in the process, like incubation of the recombinant proteins with the enzyme as well as its removal. In addition, pAE has other advantages over the pET3-His plasmid. Transformed E. coli cells with pAE produce a greater number of copies of plasmids/cell, yielding much more plasmid/miniprep. This facilitates further plasmid manipulation for subcloning, sequencing and, in theory, a higher gene dosage that could result in a larger amount of recombinant protein expression, although this was not observed in the case of Fc of tetanus toxin. Successful expression of several other proteins was also achieved using this plasmid (14-16). Furthermore, pAE possesses a multiple cloning site after the His-tag region that facilitates subcloning of the desired insert. All of these features make the pAE plasmid a very useful expression vector for E. coli.
Plasmid yield from cultures of Escherichia coli transformed with pRSETA, pET-His or pAE plasmid. DNA samples were digested with BamHI. M = 1-kb DNA ladder (Life Technologies). Lane 1, pRSETA; lane 2, pET-His; lane 3, pAE.
Diagrammatic representation of the construction of the pAE plasmid. Fc is the tetanus toxin fragment C.
Multiple cloning sites of pET3-His (A), pRSETA (B) and pAE (C). The nucleotide sequences of the multiple cloning sites, the corresponding restriction enzymes and amino acids are described. The enterokinase cleavage site present in pRSETA is indicated by the arrow.
SDS-PAGE analysis of recombinant fragment C (50 kDa) expressed in Escherichia coli BL21 (DE3) after IPTG induction. Lanes 1, 3, 5, 7, 9, 11, 13 and 15, cells were transformed with pET3-His-Fc. Lanes 2, 4, 6, 8, 10, 12, 14 and 16, cells were transformed with pAE-Fc. Lanes 1 and 2, total protein extract from non-induced cultures. The samples of induced cultures were collected every 30 min from 0 h (lanes 1 and 2) to 3.5 h (lanes 3-16). M = molecular mass markers (Pharmacia). Horizontal arrows = 50 kDa. The gels were stained with 0.25% Coomassie blue in 40% ethanol/10% acetic acid and destained with a solution of 10% ethanol.
SDS-PAGE analysis of recombinant fragment C (50 kDa) purified from Escherichia coli BL21 (DE3) transformed by pET-His-Fc and pAE-Fc. Lane 1, pET-His-Fc. Lane 2, pEA-Fc.M = molecular mass markers (Pharmacia). Eight micrograms of purified proteins was loaded onto the gels. The gels were stained with 0.25% Coomassie blue in 40% ethanol/10% acetic acid and destained with a solution of 10% ethanol.
We thank Fátima A.M. de Oliveira, Salete Vargas, Ingrid R.S. Pimenta, and Érika Regazoli for excellent technical assistance.
Correspondence and Footnotes

Address for correspondence: P.L. Ho, Centro de Biotecnologia, Instituto Butantan, Av. Vital Brasil, 1500, 05503-900, São Paulo, SP, Brasil. Fax: +55-11-3726-1505. E-mail: hoplee@butantan.gov.br
Research supported by FAPESP, CNPq and Fundação Butantan. Received August 13, 2003. Accepted May 20, 2004.
- 1. Studier FW, Rosenberg AH, Dunn JJ & Dubendorff JW (1990). Use of T7 RNA polymerase to direct expression of cloned genes. Methods in Enzymology, 185: 60-89.
- 2. Oliveira MLS, Neto JC, Krieger JE, Raw I & Ho PL (2001). Site-directed mutagenesis of bovine FGF-2 cDNA allows the production of the human-form of FGF-2 in Escherichia coli Biotechnology Letters, 23: 1151-1157.
- 3. Crowe J, Dobeli H, Gentz R, Hochuli E, Stuber D & Henco K (1994). 6xHis-Ni-NTA chromatography as a superior technique in recombinant protein expression/purification. Methods in Enzymology, 31: 271-387.
- 4. Choi JW, Ra KS & Lee YS (1999). Enhancement of bovine growth hormone gene expression by increasing the plasmid copy number. Biotechnology Letters, 21: 1-5.
- 5. Chen BP & Hai T (1994). Expression vectors for affinity purification and radiolabeling of proteins using Escherichia coli as host. Gene, 139: 73-75.
- 6. Sambrook J, Fritsch EF & Maniatis T (1989). Molecular Cloning: A Laboratory Manual 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA.
- 7. Ribas AV, Ho PL, Tanizaki MM, Raw I & Nascimento ALTO (2000). High-level expression of tetanus fragment C-thioredoxin fusion protein in Escherichia coli. Biotechnology and Applied Biochemistry, 31: 91-94.
- 8. Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW, Crosa JH & Falkow S (1977). Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene, 2: 95-113.
- 9. Yanisch-Perron C, Vieira J & Messing J (1985). Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene, 33: 103-119.
- 10. Davison J (1984). Mechanism of control of DNA replication and incompatibility in ColE1-type plasmids. Gene, 28: 1-15.
- 11. Tomizawa J & Itoh T (1981). Plasmid ColE1 incompatibility determined by interaction of RNA I with primer transcript. Proceedings of the National Academy of Sciences, USA, 78: 6096-6100.
- 12. Cesareni G, Muesing MA & Polisky B (1982). Control of ColE1 DNA replication: the rop gene product negatively affects transcription from the replication primer promoter. Proceedings of the National Academy of Sciences, USA, 79: 6313-6317.
- 13. Enomoto S, Chen G & Berman J (1998). Vectors for expressing T7 epitope- and His6 affinity-tagged fusion proteins in S. cerevisiae Biotechniques, 24: 782-788.
- 14. Junqueira de Azevedo ILM, Farsky SHP, Oliveira MLS & Ho PL (2001). Molecular cloning and expression of a functional snake venom vascular endothelium growth factor (VEGF) from the Bothrops insularis pit viper. A new member of the VEGF family of proteins. Journal of Biological Chemistry, 276: 39836-39842.
- 15. Areas APM, Oliveira MLS, Ramos CRR, Sbrogio-Almeida ME, Raw I & Ho PL (2002). Synthesis of cholera toxin B subunit gene: cloning and expression of a functional 6XHis-tagged protein in Escherichia coli Protein Expression and Purification, 25: 481-487.
- 16. Ramos CRR, Figueiredo RCR, Pertinhez TA, Vilar MM, Nascimento ALTO, Tendler M, Raw I, Spisni A & Ho PL (2003). Gene structure and M20T polymorphism of the Schistosoma mansoni Sm14 fatty acid-binding protein: Molecular, functional and immunoprotection analysis. Journal of Biological Chemistry, 278: 12745-12751.
Acknowledgments
Publication Dates
-
Publication in this collection
20 July 2004 -
Date of issue
Aug 2004
History
-
Accepted
20 May 2004 -
Received
13 Aug 2003