Abstract
Torque teno virus (TTV) is a circular, single-stranded DNA virus that chronically infects healthy individuals of all ages worldwide. TTV has an extreme genetic heterogeneity which is reflected in its current classification into five main phylogenetic groups (1-5). Using specific PCR assays, it has been shown that many individuals are co-infected with TTV isolates belonging to different phylogenetic groups. Here, a multiplex PCR assay was developed, using five recombinant plasmids. Each plasmid carried an insert of different size issued from a TTV isolate belonging to a different group. The assay was able to simultaneously amplify DNAs of TTV isolates belonging to all five phylogenetic groups. Multiplex PCR was then tested satisfactorily on DNAs extracted from 55 serum samples (47 health care workers and 8 AIDS patients). All individuals but nine were infected with at least one TTV isolate. Co-infection with multiple isolates was found in 29/47 (62%) health care workers and in 8/8 (100%) AIDS patients. A number of discrepancies were observed when results obtained with three thermostable DNA polymerases were compared. For example, four TTV phylogenetic groups were detected in a particular serum sample by using one of the three DNA polymerases, whereas the other two enzymes were able to detect only three TTV groups. However, none of the three enzymes used could be broadly considered to be more efficient than the others. Despite its limitations, the assay described here constitutes a suitable tool to visualize the degree of co-infection of a given population, avoiding time-consuming experiments.
Brazil; Co-infections; Multiplex; PCR; Torque teno virus
Braz J Med Biol Res, June 2005, Volume 38(6) 853-860
A multiplex PCR assay able to simultaneously detect Torque teno virus isolates from phylogenetic groups 1 to 5
Departamento de Virologia, Instituto Oswaldo Cruz, FIOCRUZ, Rio de Janeiro, RJ, Brasil
 References
References
 Acknowledgments
Correspondence and Footnotes
Acknowledgments
Acknowledgments
Correspondence and Footnotes
Acknowledgments
Abstract
Torque teno virus (TTV) is a circular, single-stranded DNA virus that chronically infects healthy individuals of all ages worldwide. TTV has an extreme genetic heterogeneity which is reflected in its current classification into five main phylogenetic groups (1-5). Using specific PCR assays, it has been shown that many individuals are co-infected with TTV isolates belonging to different phylogenetic groups. Here, a multiplex PCR assay was developed, using five recombinant plasmids. Each plasmid carried an insert of different size issued from a TTV isolate belonging to a different group. The assay was able to simultaneously amplify DNAs of TTV isolates belonging to all five phylogenetic groups. Multiplex PCR was then tested satisfactorily on DNAs extracted from 55 serum samples (47 health care workers and 8 AIDS patients). All individuals but nine were infected with at least one TTV isolate. Co-infection with multiple isolates was found in 29/47 (62%) health care workers and in 8/8 (100%) AIDS patients. A number of discrepancies were observed when results obtained with three thermostable DNA polymerases were compared. For example, four TTV phylogenetic groups were detected in a particular serum sample by using one of the three DNA polymerases, whereas the other two enzymes were able to detect only three TTV groups. However, none of the three enzymes used could be broadly considered to be more efficient than the others. Despite its limitations, the assay described here constitutes a suitable tool to visualize the degree of co-infection of a given population, avoiding time-consuming experiments.
Key words
Brazil, Co-infections, Multiplex, PCR, Torque teno virus
Introduction
Torque teno virus (TTV) was discovered in 1997 in the blood of a Japanese patient with post-transfusional hepatitis of unknown etiology (1). TTV is a small, non-enveloped virus with a single-stranded, circular DNA genome of negative polarity, 3.4-3.9 kb in length (2). TTV has been classified recently, together with the related Torque teno mini- virus, into a new floating genus called Anellovirus (3). Despite its DNA nature, the TTV genome shows a remarkably high degree of variability. More than 30 genotypes have been ordered recently into five major phylogenetic groups (4). TTV is a ubiquitous virus, infecting more than 80% of the general population throughout the world (5-9). Co-infection of single individuals with TTV isolates belonging to one or several phylogenetic groups occurs frequently (4,10-13). Although TTV has been suggested to be a causative agent of several diseases such as hepatitis (1), gastritis (14), and acute respiratory diseases (15,16), until now this has not been demonstrated. As for human adenoviruses and papillomaviruses, porcine circoviruses, and other viruses, a possibility still exists that a specific TTV phylogenetic group (or genotype) is especially involved in the pathogenesis of a particular disease. It is therefore important to develop a method for the identification of the different TTV phylogenetic groups infecting a single individual.
In a recent study, we analyzed the distribution of TTV genomic groups 1-5 among healthy individuals as well as in hepatitis B virus carriers and human immunodeficiency virus type 1-infected patients (17). This was done by using 5 different PCR assays able to specifically amplify the TTV DNAs of each phylogenetic group. Here, a single-tube multiplex PCR assay is described to detect and simultaneously identify human TTV belonging to the 5 main phylogenetic groups. The effectiveness of the method was tested on serum samples collected from Brazilian health care workers and AIDS patients.
Material and Methods
Plasmids
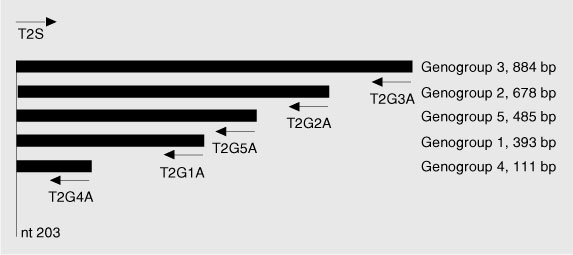
Five recombinant plasmids were used in this study. Each of them carried an insert of different size obtained from a TTV isolate belonging to a different phylogenetic group (1-5). These recombinant plasmids were obtained after cloning of PCR products sharing a common 20-bp segment at the 5' end (corresponding to the sense primer used for PCR). TTV sequences carried by the plasmids have been characterized in a previous study (17) and are available from GenBank under the following accession numbers: AY312145, phylogenetic group 1, 393 bp; AY312150, group 2, 678 bp; AY312152, group 3, 884 bp; AY312154, group 4, 111 bp; AY312158, group 5, 485 bp. Figure 1 shows the location of the insert DNAs in the TTV genome.
Serum samples and extraction of viral DNA
Forty-seven serum samples collected for a previous investigation (18) from health care workers living in Rio de Janeiro, RJ, Brazil, were employed in the present study. In addition, serum samples from eight AIDS patients were used. All eight were men who had sex with men. All samples were kept frozen until DNA extraction.
Viral DNA was extracted with phenol/chloroform after treatment of 250 µl serum with 0.5 mg/ml proteinase K in the presence of 0.2 M NaCl, 0.25% SDS, for 4 h at 37ºC. After precipitation with ethanol, the pellet was dried and resuspended in 30 µl distilled water. One microliter DNA was used in the PCR assays.
PCR assays
The PCR primers (T1S and T2S, sense; T1A, T2G1A, T2G2A, T2G3A, T2G4A and T2G5A, antisense) used in this study have been described previously (17).
i) PCR assays done with a single plasmid as the target DNA were performed with one unit of Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA) in a final volume of 50 µl. After an initial denaturation for 3 min at 94ºC, amplification occurred for 30 cycles at 94ºC for 30 s, 59ºC for 30 s, 72ºC for 1 min 15 s, followed by a final elongation at 72ºC for 7 min. A PCR sensitivity of 2,000-10,000 copies was calculated for these assays.
ii) PCR assays done with a mixture of five plasmids and different combinations of primer pairs were performed with the HotStar Taq DNA polymerase, included in the first commercially available kit especially designed for multiplex PCR (Multiplex PCR kit, QIAGEN, Hilden, Germany). This was done under the same conditions as above, except for small modifications according to manufacturer recommendations. Thus, initial DNA denaturation was at 95ºC for 15 min, and annealing and elongation steps were extended to 1 min and 30 s. No significant loss of sensitivity was observed for multiplex PCR when compared to uniplex PCR.
iii) Viral DNAs extracted from serum samples were submitted to a first round of PCR using the `universal' oligonucleotide primers T1S and T1A, that amplified a large DNA fragment covering approximately 80% of the TTV genome (17). One microliter of reaction product was used for nested multiplex PCR experiments, which were performed in the presence of six oligonucleotides in a single tube. One of the primers (T2S) was a `universal' sense primer, while the other five (T2G1A to T2G5A) were antisense primers, each of which specific for a particular TTV phylogenetic group. PCR conditions were identical to those employed with the mixture of plasmids mentioned above. Since nested assays were performed, it is likely that the PCR sensitivity was increased. All samples were tested in multiplex PCR using three different enzymes, i.e., the conventional Taq DNA polymerase (Invitrogen) and two enzymes recommended by their respective manufacturers for the use in multiplex PCR, namely AccuPrime Taq DNA polymerase (Invitrogen) and HotStar Taq DNA polymerase, the latter included in the Multiplex PCR kit (QIAGEN).
Results
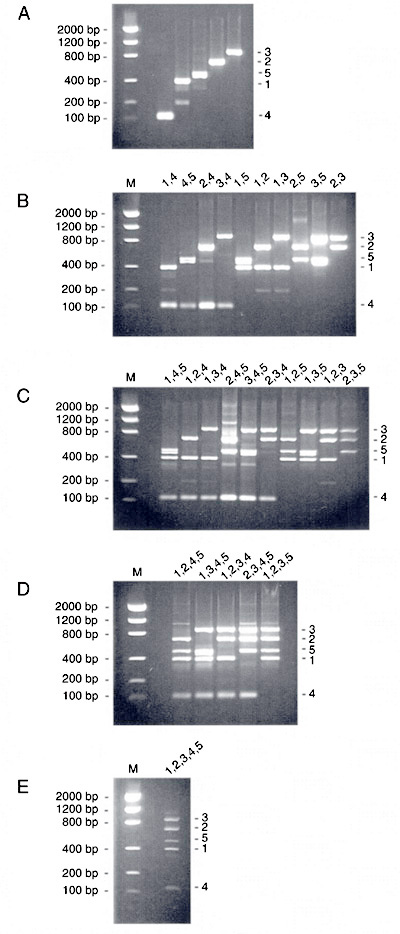
In an attempt to develop a multiplex PCR assay, a first series of experiments was performed with plasmids which were submitted to amplification in the presence of six oligonucleotide primers in a single tube. One of the primers (T2S) was a `universal' sense primer, while the other five (T2G1A to T2G5A) were antisense primers, each of which specific for a particular TTV phylogenetic group (1 to 5). This experiment was repeated five times, with a plasmid carrying a genome segment of a TTV isolate belonging to a different group each time. The results are shown in Figure 2. After agarose gel electrophoresis, DNA bands of expected sizes (393, 678, 884, 111, and 485 bp for groups 1 to 5, respectively) were obtained. Additional, non-specific bands of unexpected sizes were also observed, although most of them were barely visible.
Further experiments were performed using as DNA template a mixture of five plasmids, each carrying TTV sequences from a different phylogenetic group. This mixture was submitted to PCR amplification using the `universal' sense primer T2S associated with group-specific, antisense primers in all possible combinations. This was done to evaluate the specificity of each primer pair in a context where TTV sequences from all groups were present. Figure 3 shows the results obtained with one (A), two (B), three (C), four (D), and five (E) antisense primers present in a single tube. In general, suitable results were obtained: the number of DNA bands and their positions on the gels were as expected. In the few cases where DNA bands of unexpected sizes were observed, their intensity was lower than that of the bands corresponding to the desired fragments. There was only one exception to this rule: two adjacent DNA bands of similar intensity, about 500 bp in length, were seen in six PCR assays in which the group 5-specific antisense primer was present (Figure 3B, lanes 4,5, 1,5, and 3,5; Figure 3C, lanes 1,4,5, 3,4,5 and 1,3,5). No satisfactory explanation was found for this double band.
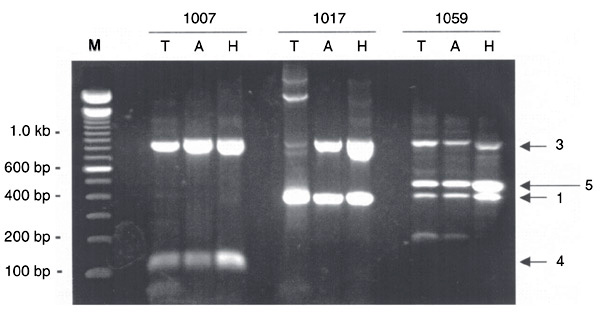
Multiplex PCR was then tested on DNAs extracted from 55 serum samples (collected from 47 health care workers and 8 AIDS patients). Each sample was tested with the conventional Taq DNA polymerase as well as with two thermostable DNA polymerases recommended for use in multiplex PCR, namely AccuPrime Taq DNA polymerase and HotStar Taq DNA polymerase. Table 1 shows the TTV phylogenetic groups detected in each sample. Thirty-eight of 47 (81%) health care workers and 8/8 (100%) AIDS patients were TTV infected. Co-infection with several TTV isolates belonging to different groups was observed in 29/47 (62%) health care workers and 8/8 (100%) AIDS patients. A number of discrepancies were observed between the experiments performed with the three enzymes, with at least one divergence in 23/55 (42%) samples. However, no system could be considered as more efficient than the others. This was because the total number of TTV groups detected with each enzyme, as summarized in Table 2, was almost identical (89, 89, and 90 for the conventional, AccuPrime, and HotStar Taq DNA polymerases, respectively). Nevertheless, as exemplified in Figure 4, the apparent DNA amounts obtained in PCR assays performed with the HotStar Taq DNA polymerase were generally higher than those obtained with the conventional Taq DNA polymerase and with the AccuPrime Taq DNA polymerase. In addition, the conventional Taq DNA polymerase generated more DNA bands of unexpected size than did the other two enzymes.
Localization of the oligonucleotides used for multiplex PCR on the Torque teno virus (TTV) genome and size of the products generated. `nt 203' refers to the position of the 5' end of the `universal' sense primer T2S (numbering according to TTV prototype TA278, Genbank accession number AB017610).
Agarose gel (2%) electrophoresis of PCR products. Each PCR assay was performed with a different plasmid and six oligonucleotide primers (one sense and five antisense). M: molecular weight markers. Numbers 1-5 at the top represent the phylogenetic groups of the Torque teno virus sequences carried by the corresponding template recombinant plasmids.
Agarose gel (2%) electrophoresis of multiplex PCR products. Each PCR assay was performed with five different plasmids (phylogenetic groups 1-5) together with one sense and one (A), two (B), three (C), four (D), or five (E) antisense oligonucleotide primers. M: Molecular weight markers. Numbers at the top of each gel indicate the Torque teno virus groups from which the pair(s) of primers added to the corresponding assays were designed.
Typical experiment showing an agarose gel (2%) electrophoresis of pentaplex PCR products derived from three serum samples (1007, 1017, and 1059). Amplification was performed with the conventional Taq DNA polymerase (T), AccuPrime Taq DNA polymerase (A) and HotStar Taq DNA polymerase (H). Numbers on the right side of the gel indicate Torque teno virus phylogenetic groups. M = molecular weight markers.
Detection of Torque teno virus genomic groups 1 to 5 by multiplex PCR in sera of health care workers and AIDS patients.
Number of Torque teno virus phylogenetic groups detected with three different thermostable DNA polymerases in multiplex PCR (55 samples analyzed).
Discussion
Concomitantly with the progressive characterization of Anellovirus genus members, the improvement of TTV PCR assays, which were initially unable to detect a wide range of genotypes, has led to increasing measurements of TTV prevalence in various populations (6,19,20). Much progress in TTV detection has been done since then, with the use of primers located in a short, highly conserved genomic region (15,21,22).
In the present study, we used plasmids carrying different TTV sequences to develop a multiplex PCR assay able to amplify in a single tube DNAs of TTV isolates belonging to all five phylogenetic groups. Multiplex PCR was then tested on DNAs extracted from 55 serum samples (47 health care workers and 8 AIDS patients). The system allowed us to detect numerous co-infections of single individuals with several TTV isolates. More precisely, 29/47 (62%) health care workers and 8/8 (100%) AIDS patients were found to be co-infected with TTV isolates belonging to different groups. The average number of TTV groups was higher among AIDS patients (2.54) than among health care workers (1.43). This is in accordance with previous observations based on uniplex PCR assays showing that immunosuppressed patients are more likely to be co-infected with several TTV isolates than other individuals (17,23).
Although the method was able to detect and characterize TTV isolates belonging to different phylogenetic groups in a single assay, some deficiencies such as the presence in some cases of DNA bands of unexpected sizes were noted. The origin of these bands (human, TTV or other) is unknown. Further cloning and nucleotide sequencing should be used to study the presence of hypothetical new TTV variants. Furthermore, some discrepancies were noted between results obtained by using three different thermostable DNA polymerases. Such discrepancies, which occurred despite the fact that no enzyme could be considered as more efficient than the others, might be due to low TTV loads in some serum samples or to low viral loads of particular phylogenetic groups in the sera of some individuals. Low amounts of target DNA, close to the limit of detection, produce PCR results showing low reproducibility. Among three thermostable DNA polymerases tested, the HotStar Taq DNA polymerase (QIAGEN) was the one which generated, at the same time, the highest amounts of DNA and the lowest number of non-specific bands in the multiplex PCR assay. For this reason, HotStar Taq DNA polymerase should be preferred to simultaneously detect TTV isolates belonging to different groups.
To characterize the different TTV isolates infecting a single individual may be important for epidemiological, transmission and pathogenicity studies. In the absence of a PCR assay using phylogenetic group- or genotype-specific primers, the classification of the different TTV isolates co-infecting single individuals requires the following steps: i) PCR amplification with generic oligonucleotide primers, ii) molecular cloning of the PCR products, and iii) isolation and nucleotide sequencing of a large number of clones. Despite its limitations, the assay described here constitutes a suitable tool to visualize the degree of co-infection of a given population, avoiding time-consuming experiments.
The authors thank Drs. Helena C.F. Vasconcelos for her assistance in the design of the PCR assays, and Selma A. Gomes for a critical reading of the manuscript.
Correspondence and Footnotes

Address for correspondence: C. Niel, Departamento de Virologia, Instituto Oswaldo Cruz, FIOCRUZ, Av. Brasil, 4365, 21040-900, Rio de Janeiro, RJ, Brasil. Fax: +55-21-2270-6397. E-mail: niel@ioc.fiocruz.br
Research supported by CNPq. Received October 7, 2004. Accepted March 28, 2005.
- 1. Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y & Mayumi M (1997). A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochemical and Biophysical Research Communications, 241: 92-97.
- 2. Miyata H, Tsunoda H, Kazi A, Yamada A, Khan MA, Murakami J, Kamahora T, Shiraki K & Hino S (1999). Identification of a novel GC-rich 113-nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus. Journal of Virology, 73: 3582-3586.
- 3. Biagini P, Todd D, Bendinelli M et al. (2005). Anellovirus In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U & Ball LA (Editors), Virus Taxonomy VIIIth Report of the ICTV. Elsevier/Academic Press, London, UK, 335-341.
- 4. Peng YH, Nishizawa T, Takahashi M, Ishikawa T, Yoshikawa A & Okamoto H (2002). Analysis of the complete genomes of thirteen TT virus variants classifiable into the fourth and fifth genetic groups, isolated from viremic infants. Archives of Virology, 147: 21-41.
- 5. Prescott LE & Simmonds P (1998). Global distribution of transfusion-transmitted virus. New England Journal of Medicine, 339: 776-777.
- 6. Takahashi K, Hoshino H, Ohta Y, Yoshida N & Mishiro S (1998). Very high prevalence of TT virus (TTV) infection in general population of Japan revealed by a new set of PCR primers. Hepatology Research, 12: 233-239.
- 7. Niel C, de Oliveira JM, Ross RS, Gomes SA, Roggendorf M & Viazov S (1999). High prevalence of TT virus infection in Brazilian blood donors. Journal of Medical Virology, 57: 259-263.
- 8. Bendinelli M, Pistello M, Maggi F, Fornai C, Freer G & Vatteroni ML (2001). Molecular properties, biology, and clinical implications of TT virus, a recently identified widespread infectious agent of humans. Clinical Microbiology Reviews, 14: 98-113.
- 9. Huang LY, Oysten Jonassen T, Hungnes O & Grinde B (2001). High prevalence of TT virus-related DNA (90%) and diverse viral genotypes in Norwegian blood donors. Journal of Medical Virology, 64: 381-386.
- 10. Biagini P, Gallian P, Attoui H, Cantaloube JF, de Micco P & de Lamballerie X (1999). Determination and phylogenetic analysis of partial sequences from TT virus isolates. Journal of General Virology, 80: 419-424.
- 11. Niel C, Saback FL & Lampe E (2000). Co-infection with multiple TT virus strains belonging to different genotypes is a common event in healthy Brazilian adults. Journal of Clinical Microbiology, 38: 1926-1930.
- 12. Saback FL, Gomes SA & Niel C (2002). High frequency of mixed TT virus infections in healthy adults and children detected by a simplified heteroduplex mobility assay. Journal of Virological Methods, 101: 117-125.
- 13. Maggi F, Pifferi M, Tempestini E et al. (2003). TT virus loads and lymphocyte subpopulations in children with acute respiratory diseases. Journal of Virology, 77: 9081-9083.
- 14. Maggi F, Marchi S, Fornai C et al. (2003). Relationship of TT virus and Helicobacter pylori infections in gastric tissues of patients with gastritis. Journal of Medical Virology, 71: 160-165.
- 15. Biagini P, Charrel RN, de Micco P & de Lamballerie X (2003). Association of TT virus primary infection with rhinitis in a newborn. Clinical Infectious Diseases, 36: 128-129.
- 16. Maggi F, Pifferi M, Fornai C et al. (2003). TT virus in the nasal secretions of children with acute respiratory diseases: Relations to viremia and disease severity. Journal of Virology, 77: 2418-2425.
- 17. Devalle S & Niel C (2004). Distribution of TT virus genomic groups 1 to 5 in Brazilian blood donors, HBV carriers, and HIV-1-infected patients. Journal of Medical Virology, 72: 166-173.
- 18. Saback FL, Palmer TE, Sabino RR, Carvalho SMF, Amorim LM, Gaspar AMC, Oliveira MLA, Yoshida CFT & Niel C (2001). Infection with hepatitis A and TT viruses and socioeconomic status in Rio de Janeiro, Brazil. Scandinavian Journal of Infectious Diseases, 33: 121-125.
- 19. Biagini P, Gallian P, Touinssi M, Cantaloube JF, Zapitelli J, de Lamballerie X & de Micco P (2000). High prevalence of TT virus infection in French blood donors revealed by the use of three PCR systems. Transfusion, 40: 590-595.
- 20. Vasconcelos HCF, Menezes ME & Niel C (2001). TT virus infection in children and adults who visited a general hospital in the south of Brazil for routine procedures. Memórias do Instituto Oswaldo Cruz, 96: 519-522.
- 21. Okamoto H, Takahashi M, Nishizawa T, Ukita M, Fukuda M, Tsuda F, Miyakawa Y & Mayumi M (1999). Marked genomic heterogeneity and frequent mixed infection of TT virus demonstrated by PCR with primers from coding and noncoding regions. Virology, 259: 428-436.
- 22. Okamoto H, Nishizawa T, Takahashi M, Tawara A, Peng Y, Kishimoto J & Wang Y (2001). Genomic and evolutionary characterization of TT virus (TTV) in tupaias and comparison with species-specific TTVs in humans and non-human primates. Journal of General Virology, 82: 2041-2050.
- 23. Shibayama T, Masuda G, Ajisawa A, Takahashi M, Nishizawa T, Tsuda F & Okamoto H (2001). Inverse relationship between the titre of TT virus DNA and the CD4 cell count in patients infected with HIV. AIDS, 15: 563-570.
Acknowledgments
Publication Dates
-
Publication in this collection
28 Sept 2005 -
Date of issue
June 2005
History
-
Accepted
28 Mar 2005 -
Received
07 Oct 2004