Abstract
When compared to other model organisms whose genome is sequenced, the number of mutations identified in the mouse appears extremely reduced and this situation seriously hampers our understanding of mammalian gene function(s). Another important consequence of this shortage is that a majority of human genetic diseases still await an animal model. To improve the situation, two strategies are currently used: the first makes use of embryonic stem cells, in which one can induce knockout mutations almost at will; the second consists of a genome-wide random chemical mutagenesis, followed by screening for mutant phenotypes and subsequent identification of the genetic alteration(s). Several projects are now in progress making use of one or the other of these strategies. Here, we report an original effort where we mutagenized BALB/c males, with the mutagen ethylnitrosourea. Offspring of these males were screened for dominant mutations and a three-generation breeding protocol was set to recover recessive mutations. Eleven mutations were identified (one dominant and ten recessives). Three of these mutations are new alleles (Otop1mlh, Foxn1sepe and probably rodador) at loci where mutations have already been reported, while 4 are new and original alleles (carc, eqlb, frqz, and Sacc). This result indicates that the mouse genome, as expected, is far from being saturated with mutations. More mutations would certainly be discovered using more sophisticated phenotyping protocols. Seven of the 11 new mutant alleles induced in our experiment have been localized on the genetic map as a first step towards positional cloning.
Mouse; Ethylnitrosourea; Mutation; Co-isogenic mutations; Mutagenesis
Braz J Med Biol Res, September 2006, Volume 39(9) 1217-1226
Inducing mutations in the mouse genome with the chemical mutagen ethylnitrosourea
 Correspondence and Footnotes
Correspondence and Footnotes
 S.M.G. Massironi1, B.L.F.S. Reis2, J.G. Carneiro2, L.B.S. Barbosa2, C.B. Ariza3, G.C. Santos2, J.L. Guénet4 and A.L.B. Godard2
S.M.G. Massironi1, B.L.F.S. Reis2, J.G. Carneiro2, L.B.S. Barbosa2, C.B. Ariza3, G.C. Santos2, J.L. Guénet4 and A.L.B. Godard2
1Departamento de Imunologia, Instituto de Ciências Biomédicas, Universidade de São Paulo, São Paulo, SP, Brasil 2Laboratório de Genética Animal e Humana, Departamento de Biologia Geral, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Belo Horizonte, MG, Brasil
3Disciplina de Biologia Molecular, Departamento de Bioquímica, Universidade Federal de São Paulo, São Paulo, SP, Brasil 4Département de Biologie du Développement, Institut Pasteur, Paris, France
 References
References
 Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Abstract
When compared to other model organisms whose genome is sequenced, the number of mutations identified in the mouse appears extremely reduced and this situation seriously hampers our understanding of mammalian gene function(s). Another important consequence of this shortage is that a majority of human genetic diseases still await an animal model. To improve the situation, two strategies are currently used: the first makes use of embryonic stem cells, in which one can induce knockout mutations almost at will; the second consists of a genome-wide random chemical mutagenesis, followed by screening for mutant phenotypes and subsequent identification of the genetic alteration(s). Several projects are now in progress making use of one or the other of these strategies. Here, we report an original effort where we mutagenized BALB/c males, with the mutagen ethylnitrosourea. Offspring of these males were screened for dominant mutations and a three-generation breeding protocol was set to recover recessive mutations. Eleven mutations were identified (one dominant and ten recessives). Three of these mutations are new alleles (Otop1mlh, Foxn1sepe and probably rodador) at loci where mutations have already been reported, while 4 are new and original alleles (carc, eqlb, frqz, and Sacc). This result indicates that the mouse genome, as expected, is far from being saturated with mutations. More mutations would certainly be discovered using more sophisticated phenotyping protocols. Seven of the 11 new mutant alleles induced in our experiment have been localized on the genetic map as a first step towards positional cloning.
Key words: Mouse, Ethylnitrosourea, Mutation, Co-isogenic mutations, Mutagenesis
Introduction
The publication of the nearly complete sequence of the genome of several mammalian species including man, mouse and rat must be considered a major achievement in modern biology. Comparisons of these sequences using specialized software have allowed geneticists to gather a large amount of new information concerning the structure and organization of mammalian genomes. However, if the computerized analysis of these sequences helps in the identification of genes and other important genomic sequences, it does not reveal much concerning their function(s) and this is why, as of today, functional information is available for no more than 30% of the mammalian genes. The challenge for the next decade of genomic research will certainly be to sort out and identify from the rest of the sequence the few percents (probably of the order of 5%) of the mammalian DNA with a functional activity and to assign a role to it. The only reliable way to reach this goal is to specifically address this question by experimentation. Later, with an increasing knowledge of the structure/function relationships, algorithms may be developed to expand more rapidly our knowledge of the mammalian genome.
Experiments performed with this aim are based on two strategies. The first one, termed "reverse genetics", consists of the production of targeted alterations in the genome leading to either the non-expression or mis-expression of a gene known only by its sequence. Such mutations are in general engineered in vitro by homologous recombination in embryonic stem (ES) cells, and once they have been induced, mutant genotypes are bred and their phenotypes carefully and thoroughly scrutinized in the context of the whole organism. Due to its nature, this strategy is a "top-down" approach. With the availability of ES cells this strategy has become very popular in many laboratories and several hundreds of transgenic mice have now been potentially generated worldwide. More recently, several transnational projects have been approved by funding agencies for the concerted and systematic production of at least one null (knockout) mutation in each and every gene of the mouse genome (1-3). There is no doubt that such projects, when completed, will dramatically increase our knowledge of the function of genes.
The second strategy for gene annotation is based on the observation of phenotypic variations in a given population followed by the molecular identification of the underlying genetic factor(s) after positional cloning. Since it proceeds from phenotype to genotype, this strategy is a "bottom-up" approach and is often called "forward genetics". Both strategies have been widely used in model organisms, in particular in the mouse, and both have advantages and drawbacks. The production of targeted mutations is a straightforward approach for the annotation of genes with relatively few limitations. One can consider, for example, that any DNA sequence whose function is unknown can be altered in vitro, at least theoretically, and mouse embryos can be produced with the mutation in the homozygous state to study the phenotypic effect resulting from the genetic alteration. With some refinements these engineered mutations can even be made conditional and/or tissue specific. However, the production of targeted alterations in the genome requires experience and rather sophisticated skills and equipment. More importantly, in many instances, it ends up with inconclusive phenotypes. This is the case, for example, when the inactivation of the targeted gene leads to early embryonic death or, conversely, when it leads to a very subtle or difficult to assess phenotype or even to no phenotype at all. Finally, the very small genes, and especially those with only one exon, are difficult to inactivate for practical reasons.
The second strategy, the positional cloning of mutations whose phenotype is obvious, is, in general, a tedious enterprise that requires the breeding of hundreds of mice to reduce the genetic interval harboring the mutant locus to a reasonable size, but it has the great advantage that it is always informative and requires only simple molecular techniques. In fact, one should consider that both strategies are complementary rather than alternative, although with the peculiarity that, while in vitro genetic engineering is universally applicable, positional cloning requires that mutations with a clear cut phenotype be available.
In this paper we report the conclusions of an experiment we undertook to produce new mouse mutations with the potent chemical mutagen ethylnitrosourea (ENU). We detected 11 new mutant alleles, 7 of which have been localized on the mouse genetic map as a first step towards positional cloning. Three of these mutations are new alleles at loci where at least 1 other mutant allele already existed. Among the other 8, at least 5 seem to be original and occurred at loci where no previous mutant allele had ever been reported before.
Material and Methods
Animals
Mice of the inbred strains BALB/c, C57BL/6 and NZB were obtained from the breeding facilities of the Department of Immunology, Institute of Biomedical Sciences, University of São Paulo (ICB-USP). During the experiment, these mice were bred and kept in micro-isolators inside rooms with sanitary barriers. The temperature was maintained at 21 ± 2ºC, with 12-h light and dark intervals. The mice were given commercial food (Nuvital - Nuvilab, Curitiba, PR, Brazil) and water ad libitum. Our experimental project was approved by the Ethics Committee of ICB-USP on September 14, 2000, with the registration number 29/2000.
ENU treatment
The BALB/c strain was chosen in our protocol because, according to the literature, it tolerates ENU treatment better than others and has already been successfully used in several mutagenesis experiments (for review, see Refs. 4-7). Another advantage of this strain is that its phenotypic characteristics are relatively well known (http://www.jax.org/phenome) and its genetic polymorphism in terms of microsatellite (and now single nucleotide polymorphisms) density has been extensively assayed (http://www.mmdap.org/snppanelres.html), facilitating the study of genetic linkage for the induced mutations. On the other hand, a major drawback of using an albino strain is that, because of epistasis, other coat color mutations cannot be detected.
ENU (Sigma, St. Louis, MO, USA) was dissolved in citrate phosphate buffer, pH 5. Male mice aged 8 to 10 weeks received a single intra-peritoneal injection of 200 or 250 mg/kg or multiple injections of 95 or 100 mg/kg at weekly intervals for 3 or 4 weeks. Treated mice were held in complete isolation for 1 week until carcinogenic substances had been cleared. Waste and bedding were detoxified by soaking in a concentrated bleach solution.
Breeding scheme
ENU treatment induces a sterile period of up to 7 months. After recovering fertility, the treated BALB/c males were mated with BALB/c females, and the offspring (G1 generation) were scrutinized to detect variations from the classical phenotype. To identify recessive mutations, G1 males were mated with normal BALB/c females and mated again with 4 of their (G2) female offspring. At least two progenies of these G2 females were backcrossed with their G1 father and were then observed to reach the minimum number of 10 G3 offspring. In doing so, we optimized the probability of detecting most of the ENU-induced recessive mutations segregating in the micro-pedigree by placing them in the homozygous state (Figure 1) (8). Statistical analysis indicates that, in these conditions, the non-observation, just by chance, of a mutant mouse in a cross where it is supposed to occur is £0.1 (at the 5% risk level). For those pedigrees where a phenodeviant was observed, either in G1 or G3, breeders from the main colony were set aside for detailed analysis, but in all cases the new mutation was kept in a BALB/c co-isogenic status.
Screening strategy used following ethylnitrosourea (ENU) mutagenesis in mice. BALB/c (ENU-treated) males were mated with several BALB/c females and their progeny (G1) were observed for dominant mutant phenotypes. G1 males were then mated with wild-type BALB/c females to produce the generation G2. Mating of the G2 females to their G1 father produced the G3 progeny that were also screened for recessive phenotypes. *Four G2 females were mated with their G1 father.
Mutant phenotype identification
Mutant phenotypes were identified by careful examination of G1 and G3 offspring, both for their behavior and integrity from birth to weaning. Mice were checked periodically and the accumulated information was used to help the phenotypic description of the potential mutant. A grid with several parameters being checked was used during the observation period, as shown in Table 1. The observation of a deviant phenotype in one pedigree was followed up to the adult age and confirmed by the observation of other mutant mice with similar phenotype in the same pedigree, either in G2 (dominant mutation) or in G3. Phenotypic characterization of each mutation, including histopathology, morphology and behavior, was achieved while genetic mapping was in progress. For most mutations, highly detailed phenotyping is now in progress and the results will be published later.
Crosses for genetic mapping
Mapping genes identified only by a phenotype requires several crosses to be set in which the phenotype in question segregates with the largest possible amount of genetic polymorphisms detectable at the DNA level (8). Since all our mutations were, by definition, induced in a BALB/c background we deliberately chose C57BL/6 or NZB strains as partner strains to cross with for the purpose of mapping. Choosing these two strains, which are distantly related to BALB/c, optimizes the distribution and density of genetic polymorphisms of all kind across the genome (9).
Dominant mutations were located by crossing the mutant mice observed in G1 (co-isogenic BALB/c) with mice of the partner strain followed by a backcross with the same partner strain (Figure 2A). For viable and fertile recessive mutations, mice homozygous for the mutant allele were crossed to the partner strain and then heterozygous F1 were either intercrossed or backcrossed with heterozygous BALB/c partners (Figure 2B1,B2). For homozygous inviable or infertile recessive mutations, heterozygotes for the mutant allele on the BALB/c background were crossed with the partner strain and the F1 progenies were crossed randomly. Only the progenies (roughly 1/4) in which mutant phenotypes were found segregating in F2 were retained for genotyping (Figure 2C). In the backcross progeny, all mice were informative and each one represented one meiotic event in the computation for genetic distances. In contrast, in F2, only the mice with a mutant phenotype were considered but each one of them was counted for two meiotic events.
Breeding schemes for the mapping of the ENU-induced mutations. A, Backcross for dominant mutations; B1, intercross and B2, backcross for viable and fertile recessive mutations; C, intercross for inviable and infertile mutations. In the backcross progeny, all mice are informative and each one of them represents one meiotic event in the computation for genetic distances. In contrast, in F2, only the mice with a mutant phenotype are considered but each one of them is counted as two meiotic events. F1 = first filial generation.
Detection of linkage and chromosomal assignment
The localization of loci affected by a new mutation was based on the detection of linkage disequilibrium after a complete genome scan was achieved using at least 50 DNA samples collected from the individual mutant mice and a panel of 54 carefully selected microsatellite markers (http://www.informatics.jax.org) polymorphic between the two strains involved in the crosses. These markers were evenly distributed throughout the different chromosomes of the mouse genetic map. Once linkage disequilibrium was found, more markers on the same chromosome were selected to confirm and refine the localization. At this step of our experimental protocol we never attempted to perform high resolution mapping as we plan to do in the future for those of our mutations that are considered worthy of positional cloning.
Results
Efficiency of the ENU treatments
Four types of ENU treatments were used in order to determine the optimal dose(s) and protocol to use with BALB/c males under our experimental conditions. We found that all four treatments led to a sterile period lasting 21 to 30 weeks (Table 2). We did not find any obvious difference in terms of mortality rate between the different doses of ENU administered. Splitting the dose into three shots of 95 or 100 mg/kg each administered at weekly intervals seems to be correlated with a higher level of fertility (after the sterile period) but does not reduce mortality in a significant manner. Mutants were induced in all cases as shown in Table 2.
Mortality, ethylnitrosourea (ENU)-induced sterile period, fertility, number of pedigrees established, and confirmed mutations scored in mice.
Mutants collected
Nineteen phenodeviants were observed in the 207 micro-pedigrees (G1 and G3) analyzed, out of which 11 mutations could be fixed as BALB/c co-isogenic strains. Table 3 describes the phenotypes observed and, for some of these mutations, the chromosomal locations.
Only 1 dominant mutation was recognized while all other were recessive. If we consider the number of phenodeviants observed in G1 it is likely that several other dominant mutations were lost mainly due to poor viability or infertility. The 10 recessive mutations, affecting skin or fur, gait, development, or behavior are listed in Table 3.
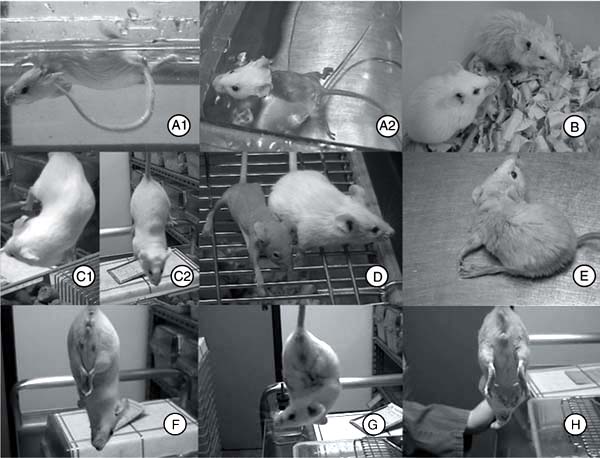
The first mutation mapped, mergulhador ("diver"; symbol - mlh), severely affects the swimming abilities of affected mice when put in a water tank (Figure 3A1 and 3A2). Under normal conditions, mlh/mlh mice exhibit head tilting but show no other behavioral abnormalities such as circling or head tossing. The locus for mlh is on chromosome 5 (Chr 5) and was identified as an allele of the mutation tilted (tlt) that affects otoconial morphogenesis without causing collateral deafness. Tilted and mergulhador are then two alleles at the Otopetrin 1 (Otop1) locus. The gene encodes a multi-transmembrane domain protein that is expressed in the macula of the developing otocyst. Both mutants carry single point mutations leading to non-conservative amino acid substitutions that affect two putative transmembrane (TM) domains (Otop1tlt is an Ala151!Glu substitution in TM3; Otop1mlh is a Leu408!Gln substitution in TM8). The existence of two mutant alleles at the Otop1 locus has facilitated the positional cloning of the tilted locus (10).
The mutation careca ("bald"; symbol - carc) induces hair abnormalities from the first coat. Mice are viable and fertile but display pronounced alteration of fur, although they have normal whiskers. The fur is sparse during the entire life of the mutant mice (Figure 3B), especially around the eyes and on the limbs and belly. Between 45 and 60 days of age, most mice lose all of their fur and become virtually naked. Histological analysis of adult skin showed an increase of skin thickness due to a large number of follicles in anagen, which is the active growth phase when the hair fiber is produced. At the same age hair follicles of control BALB/c mice were mostly in telogen, known as a resting stage. Careca has been mapped to Chr 7, between D7Mit96 (50 cM) and D7Mit174 (69 cM). In this interval of mouse Chr 7, no other mutation with a similar phenotype has been reported previously, leading us to guess that this mutation might affect a previously unknown locus.
The mutation Sacudidor de cabeça ("head shaker"; symbol - Sacc) is a dominant mutation. Mice affected by this mutation (Sacc/+) continuously shake their head but do not run in circles. The intensity of head tossing and shaking increases when the mouse is disturbed. Mice are not deaf and when stimulated by an acute sound burst (the so-called click box used for generating the Preyer reflex) their head shaking intensifies. This movement stops only when the mice are asleep. This mutation was located on Chr 15, between D15Mit100 (21 cM) and D15Mit242 (55 cM) and here, again, no other mutations with a similar phenotype have been reported.
The recessive mutation rodador ("circler"; symbol - roda) makes the mice continuously walk in circles while moving their head up and down (nodding). Here, again, stressing the mutant mice worsens the phenotype. Homozygous roda/roda mice are deaf. The roda mutation has been located on Chr 10 between D10Mit170 (29 cM) and D10Mit95 (51 cM), very close to the waltzer locus (Cadherin 23 or Otocadherin) where several other mutant alleles (actually 18) have already been reported. Given the phenotype and the linkage data we suspected roda to be a new allele at the waltzer locus and did not pursue the investigation.
Another recessive mutation discovered in our program was designated equilíbrio ("equilibrium"; symbol - eqlb) because of the abnormal motor coordination it causes. Affected mice always keep their head tilted downward (Figure 3C) and cannot stay on a rota-rod apparatus, suggesting that their cerebellum development is impaired. The equilíbrio mutant has been located on Chr 17, proximal to D17Mit113 (6 cM). No similar mutation has been reported in this region.
Mice affected by the mutation sem pelo ("furless"; symbol - sepe) never grow any fur throughout their lives (Figure 3D). In such mice, the thymus is absent and a genetic allelism test with BALB/c-nude confirmed that sepe is a new allele at the nude locus (Foxn1sepe).
Mice affected by the recessive mutation fraqueza ("weakness"; symbol - frqz) progressively lose movement coordination and die between 4 and 6 weeks of age (Figure 3E). Their lifespan can be slightly extended by enriching the cage environment and making water and food available inside the cage since the mice can no longer stand up to get them. The fraqueza mutant has been recently located on Chr 1 between D1Mit3 (11 cM) and D1Mit504 (81 cM). No spontaneous mutations with a similar phenotype have been reported in this genetic interval but several knockout alleles (in particular at the Erbb2, Ngf and Otx1 loci) exhibit a similar wasting neuromuscular syndrome. Allelism tests have not yet been done.
The recessive mutation anêmico ("anemic"; symbol - anem) is characterized by a yellowish color at birth, easily observed in a BALB/c progeny with normal and mutant littermates. Adults exhibit extremely white ears and tail. Fertility is reduced in both males and females. Preliminary phenotype characterization showed a reduction in red blood cell number and an increase in reticulocyte and leukocyte number, two symptoms specific for anemia. This mutation has not yet been assigned to a specific chromosome.
Mice affected by the recessive mutation bate palmas ("applauding"; symbol - bapa) and cruza pernas ("leg crossing"; symbol - crup) both exhibit abnormal movement of the hind limbs when held by their tail (Figure 3F,G). These reactions are suggestive of abnormal motor coordination. Phenotype characterization and mapping for these two mutations is in progress.
The recessive mutation nervoso ("nervous"; symbol - nerv) is characterized by obvious hyperactivity. Mutant mice move very rapidly when observed in their cages and shake vigorously when held by the tail. Seizures have also been observed in some mice. Phenotypic characterization and mapping are in progress.
Ethylnitrosourea (ENU)-induced mutant mice. A, an mlh/mlh mouse (A1) circles and is submerged in water, whereas a wild-type mouse (A2) swims. B, a carc/carc mouse presents sparse hair throughout its lifetime. C, an eqlb/eqlb mouse twists upwards (C1) while a wild-type mouse shows a normal position when held by its tail (C2). D, a nudesepe/nudesepe mouse presents no hair throughout its life. E, a frqz/frqz mouse loses movement coordination (45 days old). F, a bapa/bapa mouse, and G, a crup/crup mouse show abnormal movement of their hind legs when held by the tail, while a wild-type mouse (H) does not cross its legs.
Mode of inheritance, major phenotype, chromosome location, and abbreviations of ethylnitrosourea-induced mutations in mice.
Discussion
ENU is a powerful mutagen that can induce a wide variety of point mutations in the mouse genome (5,6,11). These point mutations are of various kinds, including nonsense and missense mutations and, unlike those resulting from genetic engineering in ES cells, they do not systematically nullify gene activity. Many cases have been reported where a missense mutation in a gene resulted in a less severe (or different) phenotype than would be produced by complete inactivation, and for this reason, at least, ENU mutagenesis appears to be an excellent strategy for gene annotation. Another advantage of ENU mutagenesis is that all genes in the genome are potential targets with the frequency of a hit in any one of them being (probably) proportional to the size of the gene in question. In addition, once a first mutant allele has been induced at a given locus, ENU mutagenesis can be used to efficiently generate a series of other mutant alleles at the same locus, with hits in various domains of the transcribed molecule (see Ref. 6, for details). On the contrary, gene trapping or gene targeting is possible only if the targeted genes are transcribed in the ES cells while grown in vitro, and yield mostly nonfunctional alleles. In fact, the only major drawback of ENU mutagenesis for gene annotation is that it requires comprehensive phenotyping followed by the tedious process of positional cloning.
The efficiency of the mutagenic treatment of male mice depends upon the dose and rate of administration of the chemical. With fractionation of the treatment into doses of 100 mg/kg, Hitotsumachi and colleagues (7) obtained a higher mutation rate than after administration of a single dose. In our experiment fractionation did not seem to have any obvious effect. However, splitting the ENU dose into three injections of 100 mg/kg each resulted in increased fertility.
In our experiment we identified 11 mutations, 3 of which appeared to be new alleles at loci where at least another mutant allele had already been identified (Otop1mlh, Foxn1sepe and probably rodador) and 4 are probably new and unique mutations, i.e., at loci where mutant alleles never occurred previously (carc, eqlb, frqz, and Sacc). The status for the last 4 is not yet known in enough detail to decide to which category they belong. The identification of new mutations is in agreement with previous observations made in other projects operating on a larger scale (12,13) and indicates that the mouse genome is still far from being saturated with mutations. When this is the case most of the induced mutations will fall in the first category, merely implementing the allelic series at a given locus.
At this point, it is important to emphasize that the only strategy we used to detect mutant genotypes was the observation of gross morphology, of the skin and fur, of the gait and behavior among the G1 and G3 offspring population from birth to weaning. This obviously represents a relatively superficial phenotyping excluding, for example, all the pathologies with a late onset. In the same way, recessive mutations resulting in stillbirths probably remained unnoticed and, since the mice we used were all albino and kept in highly protected environments, all mutations with an effect on coat color or resulting in immunodeficiency were also not detected. With a more comprehensive phenotyping we certainly would have detected more phenodeviants. More sophisticated methods of phenotyping such as magnetic resonance and radiology are now being developed to complement and speed up the characterization of the mutants (14).
Another important point was the choice of the BALB/c strain in our experiment. On this background we could easily recognize the ENU-induced nervoso mutation; however, when we crossed heterozygous +/nerv mice with normal (+/+) mice of the C57BL/6 strain for the purpose of mapping, we had tremendous difficulties in sorting out mutant mice from their normal counterparts in the same litter. We had the feeling that the mutation got lost in the cross while it was probably a mere background effect. In fact, we found a dramatic reduction in the proportion of nervoso (nerv/nerv) mice in the F2, leading us to believe that some genes with a strong modifying effect were segregating in this specific cross, reducing the phenotypic expression of the mutant allele in the BALB/c background. Such alterations of the phenotypic expression are commonly observed in mammalian genetics, but to explain this phenomenon in genetic terms is, in most instances, extremely difficult. In this respect, the mouse appears to be most appropriate because, in this species, it should be possible not only to identify the modifying genes by their effect on the phenotype, but also to clone them. Analyzing modifiers of the phenotypic expression will certainly be a major issue in tomorrow's medical genetics.
One of the drawbacks of using chemical mutagenesis is that, having identified an aberrant phenotype, it still requires considerable effort to identify the underlying genetic defect (15). On the other hand, chemical mutagenesis provides medical geneticists with a lot of new animal models that can be used to replicate some human diseases. Here again, the mouse mutation nervoso is a good example of a neurological syndrome that is amenable to chemical treatment and mutant mice can be used to assess the activity of new molecules even if they are not (yet) known at the molecular level. Animal models of epilepsy and of Parkinson's disease are badly needed and our nervoso mice could be of interest in this context.
The next steps of our project will be to map the most interesting of these mutations at high resolution based on their phenotype using additional crosses and markers. The information that will be gathered from the functional studies should help understand the ways genes are involved in the determinism of biological functions.
Acknowledgments
The authors are grateful to Joelma dos Santos Lima for special care with the mice.
 Address for correspondence: S.M.G. Massironi, Biotério de Experimentação, Departamento de Imunologia, ICB, USP, Av. Professor Lineu Prestes, 1730, 05508-900 São Paulo, SP, Brasil. Fax: +55-11-3091-7224. E-mail: smgmass@usp.br
Address for correspondence: S.M.G. Massironi, Biotério de Experimentação, Departamento de Imunologia, ICB, USP, Av. Professor Lineu Prestes, 1730, 05508-900 São Paulo, SP, Brasil. Fax: +55-11-3091-7224. E-mail: smgmass@usp.br
Research supported by FAPESP. Received December 20, 2005. Accepted July 26, 2006.
- 1. Schnutgen F, De-Zolt S, Van Sloun P, Hollatz M, Floss T, Hansen J, et al. Genomewide production of multipurpose alleles for the functional analysis of the mouse genome. Proc Natl Acad Sci U S A 2005; 102: 7221-7226.
- 2. Adams DJ, Biggs PJ, Cox T, Davies R, van der Weyden L, Jonkers J, et al. Mutagenic insertion and chromosome engineering resource (MICER). Nat Genet 2004; 36: 867-871.
- 3. Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, et al. The knockout mouse project. Nat Genet 2004; 36: 921-924.
- 4. Justice MJ, Carpenter DA, Favor J, Neuhauser-Klaus A, Hrabe de AM, Soewarto D, et al. Effects of ENU dosage on mouse strains. Mamm Genome 2000; 11: 484-488.
- 5. Russell WL, Kelly EM, Hunsicker PR, Bangham JW, Maddux SC, Phipps EL. Specific-locus test shows ethylnitrosourea to be the most potent mutagen in the mouse. Proc Natl Acad Sci U S A 1979; 76: 5818-5819.
- 6. Guenet JL. Chemical mutagenesis of the mouse genome: an overview. Genetica 2004; 122: 9-24.
- 7. Hitotsumachi S, Carpenter DA, Russell WL. Dose-repetition increases the mutagenic effectiveness of N-ethyl-N-nitrosourea in mouse spermatogonia. Proc Natl Acad Sci USA 1985; 82: 6619-6621.
- 8. Beier DR, Herron BJ. Genetic mapping and ENU mutagenesis. Genetica 2004; 122: 65-69.
- 9. Beck JA, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig JT, Festing MF, et al. Genealogies of mouse inbred strains. Nat Genet 2000; 24: 23-25.
- 10. Hurle B, Ignatova E, Massironi SM, Mashimo T, Rios X, Thalmann I, et al. Non-syndromic vestibular disorder with otoconial agenesis in tilted/mergulhador mice caused by mutations in otopetrin 1. Hum Mol Genet 2003; 12: 777-789.
- 11. Concepcion D, Seburn KL, Wen G, Frankel WN, Hamilton BA. Mutation rate and predicted phenotypic target sizes in ethylnitrosourea-treated mice. Genetics 2004; 168: 953-959.
- 12. Hrabe de Angelis MH, Flaswinkel H, Fuchs H, Rathkolb B, Soewarto D, Marschall S, et al. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat Genet 2000; 25: 444-447.
- 13. Nolan PM, Peters J, Strivens M, Rogers D, Hagan J, Spurr N, et al. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat Genet 2000; 25: 440-443.
- 14. Keays DA, Nolan PM. N-ethyl-N-nitrosourea mouse mutants in the dissection of behavioural and psychiatric disorders. Eur J Pharmacol 2003; 480: 205-217.
- 15. Nolan PM, Hugill A, Cox RD. ENU mutagenesis in the mouse: application to human genetic disease. Brief Funct Genomic Proteomic 2002; 1: 278-289.
Correspondence and Footnotes
Publication Dates
-
Publication in this collection
27 Feb 2008 -
Date of issue
Sept 2006
History
-
Accepted
26 July 2006 -
Received
20 Dec 2005