Abstract
Endothelins (ETs) and sarafotoxins (SRTXs) belong to a family of vasoconstrictor peptides, which regulate pigment migration and/or production in vertebrate pigment cells. The teleost Carassius auratus erythrophoroma cell line, GEM-81, and Mus musculus B16 melanocytes express rhodopsin, as well as the ET receptors, ETB and ETA, respectively. Both cell lines are photoresponsive, and respond to light with a decreased proliferation rate. For B16, the doubling time of cells kept in 14-h light (14L):10-h darkness (10D) was higher compared to 10L:14D, or to DD. The doubling time of cells kept in 10L:14D was also higher compared to DD. Using real-time PCR, we demonstrated that SRTX S6c (12-h treatment, 100 pM and 1 nM; 24-h treatment, 1 nM) and ET-1 (12-h treatment, 10 and 100 pM; 24- and 48-h treatments, 100 pM) increased rhodopsin mRNA levels in GEM-81 and B16 cells, respectively. This modulation involves protein kinase C (PKC) and the mitogen-activated protein kinase cascade in GEM-81 cells, and phospholipase C, Ca2+, calmodulin, a Ca2+/calmodulin-dependent kinase, and PKC in B16 cells. Cells were kept under constant darkness throughout the gene expression experiments. These results show that rhodopsin mRNA levels can be modulated by SRTXs/ETs in vertebrate pigment cells. It is possible that SRTX S6c binding to the ETB receptors in GEM-81 cells, and ET-1 binding to ETA receptors in B16 melanocytes, although activating diverse intracellular signaling mechanisms, mobilize transcription factors such as c-Fos, c-Jun, c-Myc, and neural retina leucine zipper protein. These activated transcription factors may be involved in the positive regulation of rhodopsin mRNA levels in these cell lines.
Rhodopsin; Sarafotoxin; Endothelin; Pigment cells; Gene expression; Signaling pathway
Braz J Med Biol Res, September 2010, Volume 43(9) 828-836
Modulation of rhodopsin gene expression and signaling mechanisms evoked by endothelins in goldfish and murine pigment cell lines
G.J.D. Lopes, C.C. Góis, L.H.R.G. Lima and  Correspondence and Footnotes
Correspondence and Footnotes
 A.M.L. Castrucci
A.M.L. Castrucci
Departamento de Fisiologia, Instituto de Biociências, Universidade de São Paulo, São Paulo, SP, Brasil
 Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Abstract
Endothelins (ETs) and sarafotoxins (SRTXs) belong to a family of vasoconstrictor peptides, which regulate pigment migration and/or production in vertebrate pigment cells. The teleost Carassius auratus erythrophoroma cell line, GEM-81, and Mus musculus B16 melanocytes express rhodopsin, as well as the ET receptors, ETB and ETA, respectively. Both cell lines are photoresponsive, and respond to light with a decreased proliferation rate. For B16, the doubling time of cells kept in 14-h light (14L):10-h darkness (10D) was higher compared to 10L:14D, or to DD. The doubling time of cells kept in 10L:14D was also higher compared to DD. Using real-time PCR, we demonstrated that SRTX S6c (12-h treatment, 100 pM and 1 nM; 24-h treatment, 1 nM) and ET-1 (12-h treatment, 10 and 100 pM; 24- and 48-h treatments, 100 pM) increased rhodopsin mRNA levels in GEM-81 and B16 cells, respectively. This modulation involves protein kinase C (PKC) and the mitogen-activated protein kinase cascade in GEM-81 cells, and phospholipase C, Ca2+, calmodulin, a Ca2+/calmodulin-dependent kinase, and PKC in B16 cells. Cells were kept under constant darkness throughout the gene expression experiments. These results show that rhodopsin mRNA levels can be modulated by SRTXs/ETs in vertebrate pigment cells. It is possible that SRTX S6c binding to the ETB receptors in GEM-81 cells, and ET-1 binding to ETA receptors in B16 melanocytes, although activating diverse intracellular signaling mechanisms, mobilize transcription factors such as c-Fos, c-Jun, c-Myc, and neural retina leucine zipper protein. These activated transcription factors may be involved in the positive regulation of rhodopsin mRNA levels in these cell lines.
Key words: Rhodopsin; Sarafotoxin; Endothelin; Pigment cells; Gene expression; Signaling pathway
Introduction
Endothelins (ETs) are 21-residue peptides, first isolated and identified in cultured porcine aortic endothelial cells (1). The three ET isoforms, ET-1, ET-2 and ET-3, share approximately 60% sequence homology with the sarafotoxins (SRTXs) (2), which are a family of peptides from the venom of Atractaspidae snakes (3).
Besides endothelial cells, many other cell types synthesize ETs, including human keratinocytes (4). The keratinocyte-derived ET-1 regulates the proliferation and melanogenesis/dendritogenesis of melanocytes in UVB- or UVA-irradiated skins (5). In cultured human melanocytes, ETs promote cell proliferation (6) and melanogenesis (6,7) through a significant increase in tyrosinase activity (7).
ETs and SRTXs also regulate pigment migration and/or production in fish pigment cells (chromatophores). In several teleostean species, ETs induce pigment aggregation in melanophores (8) and xantophores (9). The SRTX isoform S6c induces pigment aggregation in the melanophores of eight teleostean species and in Oryzias latipes xantophores (8,9). SRTXs and ETs act on the cardiovascular system of vertebrates through three ET receptor subtypes, ETA, ETB, and ETC, all belonging to the G-protein-coupled receptor superfamily (10-12). We have demonstrated that the erythrophoroma cell line of the teleost Carassius auratus, GEM-81, and B16 Mus musculus melanocytes express ETB (13) and ETA (14) receptors, respectively. ET receptors may activate a diversity of intracellular signaling pathways, including phospholipase C (PLC), phospholipase D (15) and the mitogen-activated protein kinase (MAPK) cascade (16).
Chromatophores of many poikylothermic species can directly respond to light, which triggers the dispersion or aggregation of the pigment granules along the dendritic processes of the cells, allowing the animal to display a rapid color change (17). These light-evoked responses are probably mediated by photoreceptor molecules expressed by these cells. In fact, the expression of red and green opsins has been demonstrated in light-responsive erythrophores of tilapia fins (18), and we demonstrated the presence of rhodopsin in the erythrophoroma cell line GEM-81 (19). Rhodopsin is also expressed in melan a2 cells, the immortalized murine melanocytes (20), and in B16-F10 melanoma cells (this paper), both cell lines derived from C57BL mice. The photopigment present in GEM-81 and B16 cells is probably functional, since light inhibits the proliferation of these cells (19, this paper).
Rhodopsin is the vertebrate rod photopigment. In mammals, the transcriptional regulation of rhodopsin expression involves at least two distinct regulatory regions, the rhodopsin proximal promoter region (RPPR) (21,22) and a distal region, the rhodopsin enhancer region (23). In teleost fish, the rhodopsin gene does not have introns (24), but its promoter seems to have similar sequences to those found in the RPPR of mammals and can bind conserved transcription factors (25,26).
Here we demonstrate that SRTX S6c and ET-1 modulate rhodopsin mRNA levels in the photosensitive GEM-81 and B16 cells, respectively. This modulation involves protein kinase C (PKC) and the MAPK cascade in GEM-81 cells, and PLC, Ca2+, calmodulin, a Ca2+/calmodulin-dependent kinase, and PKC in B16 cells.
Material and Methods
Cell cultures and proliferation assays
GEM-81 cells, derived from goldfish (C. auratus) cutaneous tumors, were kept in Ham's F-10 medium (Cultilab, Brazil) supplemented with 10% fetal calf serum (FCS; Nutricell, Brazil), 14.3 mM NaHCO3, 15 mM hydroxyethylpiperazineethanesulfonic acid (HEPES; Gibco, USA), 1% penicillin/streptomycin/amphotericin B (Gibco), pH 7.2, at 25°C.
Mus musculus B16 melanocytes were kept in Ham's F-10 medium (Cultilab) supplemented with 10% FCS (Cultilab), 14.3 mM NaHCO3, 15 mM HEPES (Gibco), 1% penicillin/streptomycin/amphotericin B (Gibco), pH 7.2, at 37°C and 5% CO2, in a humidified incubator.
For the proliferation experiments, B16 melanocytes were kept in constant darkness (DD) for 5 days, and then seeded (106 cells/25-cm2 flask) and kept in 10-h light:14-h darkness (10L:14D); 14-h light:10-h darkness (14L:10D), or constant darkness (DD) regimens. The medium was changed 24 h after seeding and every 48 h thereafter. Cells were harvested every 24 h with a Tyrode/EDTA solution, re-suspended in 0.5% Trypan blue-containing PBS, and counted with the aid of a hemocytometer.
Hormone assays and determination of intracellular signaling mechanisms
GEM-81 and B16 cells were seeded (3 x 106 cells/25-cm2 flask and 106 cells/25-cm2 flask, respectively) and kept in the same media as previously described, except for the fact that the FCS concentration in the medium was reduced to 2% for both cell lines, and that 1.5% dimethylsulfoxide (DMSO; Labsynth, Brazil) was added to the medium of GEM-81 cells. Cells were kept under constant darkness and the culture medium was replaced 24 and 72 h after seeding, under red light. For GEM-81 cells, DMSO was removed before the treatments described below.
For the hormone assays, the culture medium was replaced with medium containing 10 pM to 100 nM SRTX S6c (Calbiochem, USA) or 10 pM to 100 nM ET-1 (Calbiochem), for GEM-81 and B16 cells, respectively, 5 days after seeding. GEM-81 cells were treated with SRTX S6c for 6, 12, or 24 h and B16 cells were treated with ET-1 for 6, 12, 24, and 48 h. After each treatment, total RNA was extracted, followed by RT-PCR and real-time (quantitative) PCR.
To investigate the intracellular signaling mechanisms involved in the modulation of rhodopsin mRNA levels, 5 days after seeding the cells were treated for 24 h with 1 nM SRTX S6c (GEM-81 cells; Calbiochem) or 100 pM ET-1 (B16 cells; Calbiochem), in the presence or absence of intracellular signaling pathway blockers [3 µM U-73122, a PLC inhibitor (Biomol, USA); 10 µM BAPTA-AM, a calcium chelator (Research Biochemicals International, USA); 1 µM R 24571, a calmodulin antagonist (Biomol); 3 µM KN-93, a Ca2+/calmodulin-dependent kinase II inhibitor (Biomol); 3 µM Ro31-8220, a PKC inhibitor (Biomol); 10 nM GW 5074, a cRaf-1 inhibitor (Biomol); 20 µM PD-98059, a MEK inhibitor (Calbiochem); 0.53 µM 5-iodotubercidin, an ERK2 inhibitor (Biomol)]. After each treatment, total RNA was extracted, followed by RT-PCR and real-time (quantitative) PCR.
Total RNA extraction, RT-PCR, PCR, cloning, and sequencing
Total RNA was extracted with TRIzol reagent (Invitrogen, USA) and after DNase treatment (according to manufacturer instructions; turboDNA-free kit, Ambion Inc., USA), total RNA concentration was determined with a spectrophotometer (GeneQuant Pro, Biochrom Ltd., UK, or NanoDrop ND-1000 Spectrophotometer, NanoDrop, USA) and 1 µg was reverse transcribed with Superscript III enzyme according to manufacturer instructions (Invitrogen).
For PCR, the primer sequences were designed with the Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3.cgi), based on the rhodopsin sequence (accession No. NM_145383.1, M. musculus Rho) obtained from GenBank (www.ncbi.nlm.nih.gov/PubMed).
The PCR mixture contained 1 μL cDNA, 2.5 μL 10X PCR buffer (without Mg2+), 1.5 μL MgCl2 (25 mM), 4 μL dNTPs (1.25 mM) all from Invitrogen, 2 μL rhodopsin forward primer (6.4 ρmol/μL, 5'-GAGGGCCCCAATTTTTATGT-3'), 2 μL rhodopsin reverse primer (6.4 ρmol/μL, 5'-GTCGTC ATCTCCCAGTGGAT-3'), 0.3 μL AmpliTaq Gold (Applied Biosystems, USA), and nuclease-free water (Ambion). PCR was carried out in an Eppendorf thermocycler (Mastercycler Personal; Eppendorf, Germany), with the following amplification profile: 6 min at 95°C, followed by 35 cycles of 30 s at 95°C, 1 min at 60°C and 1 min at 72°C, ending with 1 cycle of 10 min at 72°C. Negative controls without template were routinely included. The size of the PCR products was verified on 1.2% agarose gel in Tris acetate buffer stained with 1% ethidium bromide, and DNA was visualized with a UV-transilluminator. The images were digitized with the Gel Pro Image system (Fotodyne Inc., USA).
The band corresponding to the expected size was cut from the gel, and the fragment was extracted by centrifugation at 1000 g for 10 min in 50-μL filter tips (Beveled filter tips; USA Scientific Inc., USA) for agarose removal, and through Millipore (USA) columns for salt removal. Using a TOPO TA Cloning kit (Invitrogen), the plasmid linked to the PCR product was inserted into One Shot competent cells. Colonies containing the insert were grown overnight and submitted to DNA purification with the Mini-Prep kit (Promega, USA). After DNA quantification with a spectrophotometer, a new PCR was performed for sequencing with 400-500 ηg plasmid, 6.4 ρmol T7 primer, 4 μL Big Dye (Applied Biosystems), with the final volume adjusted to 20 μL with distilled water. The total volume was passed through columns (Edge Biosystems, USA), evaporated and re-suspended in 20 μL ABI buffer (Applied Biosystems) for sequencing.
The sequences obtained with the plasmid fragment subtracted were submitted to comparison at the National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health site (www.ncbi.nlm.nih.gov), in the BLAST program, and the nucleotide sequence was translated into an amino acid sequence to confirm the identity of the mRNA.
Quantitative PCR
The products of reverse transcription were used for real-time (quantitative) PCR using a pair of specific primers for C. auratus rhodopsin (forward 5'-CGCGTACATGTTCTTCTTG ATTAT-3', reverse 5'-TGCTCGATGGTGACGTACAGA-3'; GenBank accession No. L11863.1, C. auratus CRAROD) or M. musculus rhodopsin (forward 5'-TGCCACACTTGGAGG TGAAA-3', reverse 5'-ACCACGTAGCGCTCAATGG-3'; GenBank accession No. NM_145383.1, M. musculus Rho). The 18S ribosomal RNA (forward primer 5'-CGGCTACC ACATCCAAGGAA-3', reverse primer 5'-GCTGGAATTACC GCGGCT-3') was used as a control to normalize the rhodopsin values. The primers were designed by the Primer Express program (Applied Biosystems).
The reaction mixtures were prepared with the primers (300 ηM for rhodopsin, 50 ηM for 18S RNA), iQ SYBR Green Supermix 2X (BioRad Laboratories, USA) or SYBR GreenER qPCR SuperMix for iCycler 2X (Invitrogen) and DNaseRNase-free water (Ambion Inc.). The reactions were carried out in the iCycler thermocycler (BioRad Laboratories), under the following conditions: 3 min at 95°C, followed by 40 cycles of 10 s at 95°C and 45 s at 55°C, 1 min at 95°C, 1 min at 55°C, 80 cycles of 10 s at 55°C, with a gradual rise of 0.5°C, for the iQ SYBR Green Supermix 2X. When the mix was purchased from Invitrogen (SYBR GreenER qPCR SuperMix for iCycler 2X), the manufacturer protocol was followed.
The results were transformed, with the rhodopsin mRNA levels of the control group being 100%.
Statistical analysis
For the proliferation and the real-time PCR assays, the data were compared by one-way analysis of variance (ANOVA), followed by the Tukey test. The differences were considered to be significant when P < 0.05.
Results
Proliferation and rhodopsin expression
To investigate the photosensitivity of B16 melanocytes, cells were kept in constant darkness (DD) for 5 days and then transferred to different photoperiodic regimens: DD, 14L:10D and 10L:14D (Figure 1). There was a significant difference in the doubling times of cells kept in 14L:10D (58.3 ± 0.872 h; R2 = 0.87) compared to 10L:14D (55.1 ± 0.689 h; R2 = 0.88, P < 0.05), or to DD (45.6 ± 0.604 h; R2 = 0.96, P < 0.01). The doubling time of cells kept in 10L:14D was also higher (P < 0.01) when compared to cells kept in DD. Viable and non-viable cells, as well as loose cells in the medium, were counted, and the results indicated that loss of adherence or cell death could not account for the decrease in cell number.
Considering that B16 murine melanocytes exhibit the above response to light, we investigated the expression of the known photopigment, rhodopsin, by these cells. Rhodopsin mRNA was confirmed by RT-PCR followed by PCR, cloning, and sequencing (data not shown).
Rhodopsin mRNA levels
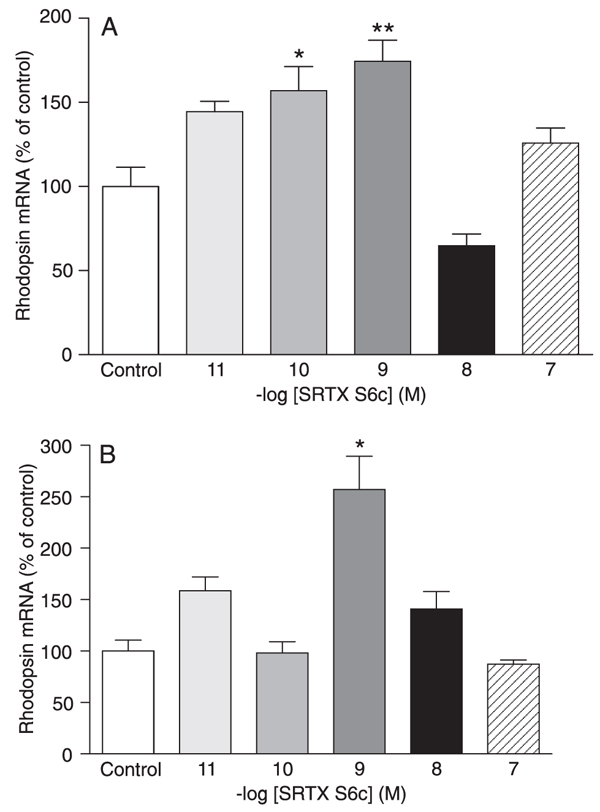
In DMSO-differentiated C. auratus GEM-81 cells, treatment with 10 pM to 100 nM SRTX S6c for 6 h did not alter rhodopsin mRNA levels (data not shown). When cells were submitted to a 12-h SRTX S6c treatment, a significant increase in rhodopsin mRNA levels was observed at the concentrations of 100 pM and 1 nM (57 and 74%, P < 0.05 and P < 0.01, respectively; Figure 2A). Treatment with SRTX S6c for 24 h promoted a significant 157% increase in rhodopsin mRNA levels only at the concentration of 1 nM (P < 0.001; Figure 2B).
In B16 cells, treatment with 10 pM to 100 nM ET-1 for 6 h did not affect rhodopsin mRNA levels (data not shown). Treatment with ET-1 for 12 h promoted a significant increase in rhodopsin mRNA levels at the concentrations of 10 and 100 pM (90 and 124%, P < 0.05 and P < 0.001, respectively; Figure 3A). In the 24- and 48-h treatments, only 100 pM ET-1 significantly increased rhodopsin mRNA levels (255.5 and 320.5%, respectively, P < 0.01; Figure 3B,C).
Intracellular signaling mechanisms
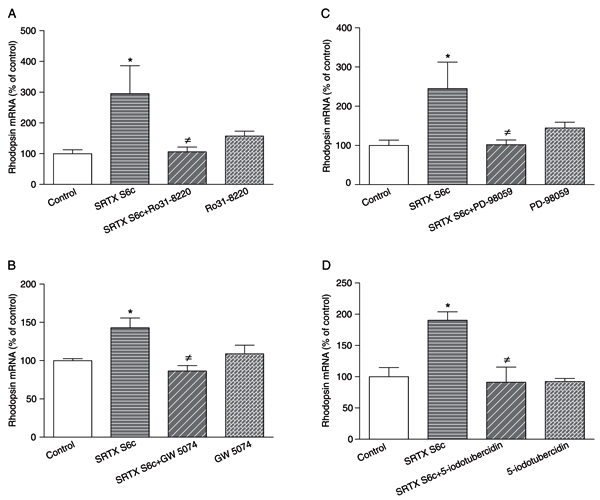
The presence of a PLC inhibitor (U-73122, 3 µM), a calcium chelator (BAPTA-AM, 10 µM), a calmodulin antagonist (R 24571, 1 µM) or a Ca2+/calmodulin-dependent kinase II inhibitor (KN-93, 3 µM) did not affect the increase in rhodopsin mRNA levels promoted by the 24-h treatment with 1 nM SRTX S6c of GEM-81 cells (data not shown). However, when GEM-81 cells were treated with 1 nM SRTX for 24 h in the presence of a PKC inhibitor (Ro31-8220, 3 µM), a cRaf-1 inhibitor (GW 5074, 10 nM), a MEK inhibitor (PD-98059, 20 µM) or an ERK2 inhibitor (5-iodotubercidin, 0.53 µM), the increase in rhodopsin mRNA levels promoted by SRTX S6c was significantly abolished (Figure 4A-D).
In B16 melanocytes, the increase in rhodopsin mRNA levels promoted by the 24-h treatment with 100 pM ET-1 was significantly blocked by a PLC inhibitor (U-73122, 3 µM), a calcium chelator (BAPTA-AM, 10 µM), a calmodulin antagonist (R 24571, 1 µM), a Ca2+/calmodulin-dependent kinase II inhibitor (KN-93, 3 µM), or a PKC inhibitor (Ro31-8220, 3 µM; Figure 5A-E).
In B16 melanocytes, the presence of a cRaf-1 inhibitor (GW 5074, 10 nM), a MEK inhibitor (PD-98059, 20 µM) or an ERK2 inhibitor (5-iodotubercidin, 0.53 µM) did not affect the increase in rhodopsin mRNA levels promoted by the 24-h treatment with 100 pM ET-1 (data not shown).
Proliferation of B16 cells submitted to different photoperiod regimens (10-h light:14-h darkness (10L:14D); 14-h light:10-h darkness (14L:10D); or constant darkness (DD). Each point is the mean (N = 5 flasks) ± SEM (if not shown, SEM are smaller than symbols). Differences were tested by one-way ANOVA followed by the Tukey test. The cell number was counted daily for 9 days. A significant difference was found in the doubling time of cells kept in 14L:10D (58.3 ± 0.872 h; R2 = 0.87) compared to 10L:14D (55.1 ± 0.689 h; R2 = 0.88, P < 0.05), or to DD (45.6 ± 0.604 h; R2 = 0.96, P < 0.01). The doubling time of cells kept in 10L:14D was also significantly higher (P < 0.01) when compared to cells kept in DD.
Rhodopsin mRNA levels of DMSO-differentiated Carassius auratus GEM-81 erythrophoroma cells treated with 10 pM to 100 nM sarafotoxin (SRTX) S6c, kept under constant darkness. Data are reported as means ± SEM. Differences were tested by one-way ANOVA followed by the Tukey test. A, Cells were treated for 12 h (N = 3-5 flasks), *P < 0.05 vs control, **P < 0.01 vs control. B, Cells were treated for 24 h (N = 3-6 flasks), *P < 0.001 vs control.
Rhodopsin mRNA levels of Mus musculus B16 melanocytes treated with 10 pM to 100 nM endothelin-1 (ET-1), kept under constant darkness. Data are reported as means ± SEM. Differences were tested by one-way ANOVA followed by the Tukey test. A, Cells were treated for 12 h (N = 6 flasks), *P < 0.05 vs control, **P < 0.001 vs control. B, Cells were treated for 24 h (N = 3-4 flasks), *P < 0.01 vs control. C, Cells were treated for 48 h (N = 3-5 flasks), *P < 0.01 vs control.
Rhodopsin mRNA levels of DMSO-differentiated Carassius auratus GEM-81 erythrophoroma cells treated with 1 nM sarafotoxin (SRTX) S6c in the absence or presence of various blockers for 24 h, kept under constant darkness. Data are reported as means ± SEM. Differences were tested by one-way ANOVA, followed by the Tukey test. A, 3 µM Ro31-8220 (N = 6-8 flasks), *P < 0.05 vs control, ≠P < 0.05 vs SRTX S6c. B, 10 nM GW 5074 (N = 3-5 flasks), *P < 0.05 vs control, ≠P < 0.01 vs SRTX S6c. C, 20 µM PD-98059 (N = 4-7 flasks), *P < 0.05 vs control, ≠P < 0.05 vs SRTX S6c. D, 0.53 µM 5-iodotubercidin (N = 3-6 flasks), *P < 0.05 vs control, ≠P < 0.05 vs SRTX S6c.
Rhodopsin mRNA levels of Mus musculus B16 melanocytes treated with 100 pM endothelin-1 (ET-1) in the absence or presence of various blockers for 24 h, kept under constant darkness. Data are reported as means ± SEM. Differences were tested by one-way ANOVA, followed by the Tukey test. A, 3 µM U-73122 (N = 5-6 flasks), *P < 0.05 vs control, ≠P < 0.001 vs ET-1. B, 10 µM BAPTA-AM (N = 4-7 flasks), *P < 0.01 vs control, ≠P < 0.05 vs ET-1. C, 1 µM R 24571 (N = 5-7 flasks), *P < 0.001 vs control, ≠P < 0.001 vs ET-1. D, 3 µM KN-93 (N = 4-9 flasks), *P < 0.01 vs control, ≠P < 0.001 vs ET-1. E, 3 µM Ro31-8220 (N = 4-8 flasks), *P < 0.01 vs control, ≠P < 0.001 vs ET-1.
Discussion
The effects of sarafotoxin and endothelin on teleost and human pigment cells, and on gene expression in different cell types have been described. However, it is not known whether these agonists can modulate photopigment gene expression in pigment cells. The photopigment rhodopsin is expressed by the teleost C. auratus erythrophoroma cell line, GEM-81 (19), and B16 M. musculus melanocytes (this paper). Here we demonstrated that rhodopsin mRNA levels can be modulated by SRTX S6c and ET-1 in GEM-81 and B16 cells, respectively, in a time- and dose-dependent manner. Imokawa et al. (7) reported that the incubation of human melanocytes with 10 nM ET-1 for 24 h increases tyrosinase activity and melanogenesis, and the levels of tyrosinase and tyrosinase-related protein-1 transcripts. Our results show that ET-1 evoked an increase in rhodopsin mRNA levels in B16 cells at a lower concentration (100 pM) than reported above. We found that the ability of ET-1 to increase gene expression was already observed after 12 h of treatment, and extended to 48 h. The fact that after 12 h, high concentrations of ET-1 (1 to 100 nM) or SRTX S6c (10 to 100 nM) did not evoke any response in either B16 melanocytes or GEM-81 cells strongly suggests a dose-related desensitization. SRTX S6c (100 pM) was able to positively modulate the mRNA levels of rhodopsin in GEM-81 cells with 12 h of treatment, and no effect was observed with 24 h, suggesting the occurrence of temporal desensitization. In B16 melanocytes, the occurrence of temporal desensitization is suggested by the fact that the positive effect of 10 pM ET-1 on rhodopsin mRNA levels was observed only after 12 h of treatment, and no effect was observed after 24 or 48 h.
ETA and ETB receptors can undergo rapid agonist-promoted desensitization. After internalization, both receptors are transported to early endosomes. However, while ETA receptors are recycled through the pericentriolar recycling compartment, ETB receptors are directed to lysosomes for degradation (27). The presence of high concentrations or the maintenance of low concentrations of SRTX S6c or ET-1 in our systems may have promoted the internalization of the receptors expressed by GEM-81 and B16 cells, resulting in the time- and dose-dependent desensitization observed in these cell lines.
ET-1 positively regulates the transcription of several genes in a variety of cell types, such as cardiomyocytes (28), vascular smooth muscle cells (VSMCs) (29) and rat-1 fibroblasts (30). ET-1 stimulates the transcription of the c-fos and c-myc genes in VSMCs (31). In rat fibroblasts, ET-1 rapidly induces an increase in mRNA levels of five members of the fos/jun gene family (c-fos, fos-B, fos-i, c-jun, and jun-B). Because the intracellular Ca2+ chelator, 1,2-bis(2-aminophenoxy) ethane-N-N-N'-N'-tetraacetic acid, blocked this response, increased [Ca2+]i must be directly involved in the induction of fos/jun genes by ET-1 in these cells (32). In VSMCs, the ET-1-induced immediate early c-fos gene expression is prevented by inhibitors of PKC (33). ET-1 has been reported to activate (phosphorylate) MAPK in cultured VSMCs (34).
In teleostean fish, there is evidence that pigment aggregation promoted by ETs involves PLC activation, inositol trisphosphate (IP3) production and elevation of [Ca2+]i (17). However, our data demonstrated that in GEM-81 cells, the modulation of rhodopsin mRNA levels by SRTX S6c involves the activation of a PKC and the MAPK cascade. Our results also showed that a PLC, Ca2+ as a second messenger, calmodulin or a Ca2+/calmodulin-dependent kinase are not involved in this effect. The lack of participation of Ca2+ as a second messenger suggests the involvement of calcium-independent PKC. The MAPK cascade is related to cell proliferation, differentiation and death mechanisms (35). Components of this cascade are phosphorylated and activated in response to several stimuli, promoting gene expression (36). ET-1, through ETA and ETB receptors, activates the MAPK cascade by ERK1/2 phosphorylation in many cell types, such as fibroblasts (37).
Our data showed that in B16 melanocytes the modulation of rhodopsin mRNA levels by ET-1 results from the activation of PKC and PLC, Ca2+ as a second messenger, calmodulin, and a Ca2+/calmodulin-dependent kinase. This modulation does not involve the MAPK cascade. Similarly, in cultured human melanocytes, ET-1 promotes cell proliferation and melanogenesis through PLC activation, a rise in IP3 (6) and [Ca2+]i levels (6,38) and PKC activation (39). Mitogens, which stimulate phosphoinositide hydrolysis and activate PKC, induce the expression of proto-oncogenes, including c-fos and c-myc (39).
The regulation of rhodopsin gene expression seems to have been conserved throughout evolution, and teleost fish and mammals exhibit similar transcription binding sequences in the promoter region of this gene (25,26). It is possible that SRTX S6c binding to ETB receptors in GEM-81 cells, and ET-1 binding to ETA receptors in B16 melanocytes, although activating diverse intracellular signaling mechanisms, mobilize transcription factors such as c-Fos, c-Jun, c-Myc and neural retina leucine zipper protein. These activated transcription factors could bind the neural retina leucine zipper response element in the RPPR and interact with the cone-rod homeobox, thus activating the rhodopsin promoter and positively regulating rhodopsin mRNA levels in these cell lines.
References
1. Yanagisawa M, Kurihara H, Kimura S, Tomobe Y, Kobayashi M, Mitsui Y, et al. A novel potent vasoconstrictor peptide produced by vascular endothelial cells. Nature 1988; 332: 411-415.
2. Ducancel F. The sarafotoxins. Toxicon 2002; 40: 1541-1545.
3. Kochva E, Viljoen CC, Botes DP. A new type of toxin in the venom of snakes of the genus Atractaspis (Atractaspidinae). Toxicon 1982; 20: 581-592.
4. Yohn JJ, Morelli JG, Walchak SJ, Rundell KB, Norris DA, Zamora MR. Cultured human keratinocytes synthesize and secrete endothelin-1. J Invest Dermatol 1993; 100: 23-26.
5. Hirobe T. Role of keratinocyte-derived factors involved in regulating the proliferation and differentiation of mammalian epidermal melanocytes. Pigment Cell Res 2005; 18: 2-12.
6. Yada Y, Higuchi K, Imokawa G. Effects of endothelins on signal transduction and proliferation in human melanocytes. J Biol Chem 1991; 266: 18352-18357.
7. Imokawa G, Miyagishi M, Yada Y. Endothelin-1 as a new melanogen: coordinated expression of its gene and the tyrosinase gene in UVB-exposed human epidermis. J Invest Dermatol 1995; 105: 32-37.
8. Hayashi H, Nakamura S, Fujii R. The endothelin receptors that mediate aggregation of pigment in fish melanophores. Comp Biochem Physiol B 1996; 115: 143-152.
9. Murata N, Fujii R. Pigment-aggregating action of endothelins on medaka xantophores. Zool Sci 2000; 17: 853-862.
10. Arai H, Hori S, Aramori I, Ohkubo H, Nakanishi S. Cloning and expression of a cDNA encoding an endothelin receptor. Nature 1990; 348: 730-732.
11. Sakurai T, Yanagisawa M, Takuwa Y, Miyazaki H, Kimura S, Goto K, et al. Cloning of a cDNA encoding a non-isopeptide-selective subtype of the endothelin receptor. Nature 1990; 348: 732-735.
12. Karne S, Jayawickreme CK, Lerner MR. Cloning and characterization of an endothelin-3 specific receptor (ETC receptor) from Xenopus laevis dermal melanophores. J Biol Chem 1993; 268: 19126-19133.
13. Filadelfi AM, Ramanzini GC, Visconti MA, Castrucci AM. The endothelin/sarafotoxin-induced increase of the proliferation of undifferentiated and DMSO-differentiated GEM-81 goldfish erythrophoroma cells is mediated by ETB receptors. Pigment Cell Res 2004; 17: 480-487.
14. Scarparo AC, Isoldi MC, de Lima LH, Visconti MA, Castrucci AM. Expression of endothelin receptors in frog, chicken, mouse and human pigment cells. Comp Biochem Physiol A Mol Integr Physiol 2007; 147: 640-646.
15. Ambar I, Sokolovsky M. Endothelin receptors stimulate both phospholipase C and phospholipase D activities in different cell lines. Eur J Pharmacol 1993; 245: 31-41.
16. Kasuya Y, Abe Y, Hama H, Sakurai T, Asada S, Masaki T, et al. Endothelin-1 activates mitogen-activated protein kinases through two independent signalling pathways in rat astrocytes. Biochem Biophys Res Commun 1994; 204: 1325-1333.
17. Fujii R. The regulation of motile activity in fish chromatophores. Pigment Cell Res 2000; 13: 300-319.
18. Ban E, Kasai A, Sato M, Yokozeki A, Hisatomi O, Oshima N. The signaling pathway in photoresponses that may be mediated by visual pigments in erythrophores of Nile tilapia. Pigment Cell Res 2005; 18: 360-369.
19. Im LH, Isoldi MC, Scarparo AC, Visconti MA, Castrucci AM. Rhythmic expression, light entrainment and alpha-MSH modulation of rhodopsin mRNA in a teleost pigment cell line. Comp Biochem Physiol A Mol Integr Physiol 2007; 147: 691-696.
20. Miyashita Y, Moriya T, Kubota T, Yamada K, Asami K. Expression of opsin molecule in cultured murine melanocyte. J Investig Dermatol Symp Proc 2001; 6: 54-57.
21. Chen S, Zack DJ. Ret 4, a positive acting rhodopsin regulatory element identified using a bovine retina in vitro transcription system. J Biol Chem 1996; 271: 28549-28557.
22. Zack DJ, Bennett J, Wang Y, Davenport C, Klaunberg B, Gearhart J, et al. Unusual topography of bovine rhodopsin promoter-lacZ fusion gene expression in transgenic mouse retinas. Neuron 1991; 6: 187-199.
23. Nie Z, Chen S, Kumar R, Zack DJ. RER, an evolutionarily conserved sequence upstream of the rhodopsin gene, has enhancer activity. J Biol Chem 1996; 271: 2667-2675.
24. Fitzgibbon J, Hope A, Slobodyanyuk SJ, Bellingham J, Bowmaker JK, Hunt DM. The rhodopsin-encoding gene of bony fish lacks introns. Gene 1995; 164: 273-277.
25. Kawamura S, Takeshita K, Tsujimura T, Kasagi S, Matsumoto Y. Evolutionarily conserved and divergent regulatory sequences in the fish rod opsin promoter. Comp Biochem Physiol B Biochem Mol Biol 2005; 141: 391-399.
26. Liu Y, Shen Y, Rest JS, Raymond PA, Zack DJ. Isolation and characterization of a zebrafish homologue of the cone rod homeobox gene. Invest Ophthalmol Vis Sci 2001; 42: 481-487.
27. Bremnes T, Paasche JD, Mehlum A, Sandberg C, Bremnes B, Attramadal H. Regulation and intracellular trafficking pathways of the endothelin receptors. J Biol Chem 2000; 275: 17596-17604.
28. Cheng TH, Shih NL, Chen CH, Lin H, Liu JC, Chao HH, et al. Role of mitogen-activated protein kinase pathway in reactive oxygen species-mediated endothelin-1-induced beta-myosin heavy chain gene expression and cardiomyocyte hypertrophy. J Biomed Sci 2005; 12: 123-133.
29. Rodriguez-Vita J, Ruiz-Ortega M, Ruperez M, Esteban V, Sanchez-Lopez E, Plaza JJ, et al. Endothelin-1, via ETA receptor and independently of transforming growth factor-beta, increases the connective tissue growth factor in vascular smooth muscle cells. Circ Res 2005; 97: 125-134.
30. Muldoon LL, Rodland KD, Forsythe ML, Magun BE. Stimulation of phosphatidylinositol hydrolysis, diacylglycerol release, and gene expression in response to endothelin, a potent new agonist for fibroblasts and smooth muscle cells. J Biol Chem 1989; 264: 8529-8536.
31. Komuro I, Kurihara H, Sugiyama T, Yoshizumi M, Takaku F, Yazaki Y. Endothelin stimulates c-fos and c-myc expression and proliferation of vascular smooth muscle cells. FEBS Lett 1988; 238: 249-252.
32. Pribnow D, Muldoon LL, Fajardo M, Theodor L, Chen LY, Magun BE. Endothelin induces transcription of fos/jun family genes: a prominent role for calcium ion. Mol Endocrinol 1992; 6: 1003-1012.
33. Bobik A, Grooms A, Millar JA, Mitchell A, Grinpukel S. Growth factor activity of endothelin on vascular smooth muscle. Am J Physiol 1990; 258: C408-C415.
34. Koide M, Kawahara Y, Tsuda T, Ishida Y, Shii K, Yokoyama M. Endothelin-1 stimulates tyrosine phosphorylation and the activities of two mitogen-activated protein kinases in cultured vascular smooth muscle cells. J Hypertens 1992; 10: 1173-1182.
35. Stork PJ, Schmitt JM. Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol 2002; 12: 258-266.
36. Kolch W. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem J 2000; 351 (Part 2): 289-305.
37. Naicker S, Bhoola KD. Endothelins: vasoactive modulators of renal function in health and disease. Pharmacol Ther 2001; 90: 61-88.
38. Imokawa G, Yada Y, Kimura M. Signalling mechanisms of endothelin-induced mitogenesis and melanogenesis in human melanocytes. Biochem J 1996; 314 (Part 1): 305-312.
39. Palumbo AP, Rossino P, Comoglio PM. Bombesin stimulation of c-fos and c-myc gene expression in cultures of Swiss 3T3 cells. Exp Cell Res 1986; 167: 276-280.
Acknowledgments
We are thankful to Márcio de Patto Lima for excellent technical assistance. Research supported by FAPESP and CNPq. G.J.D. Lopes and L.H.R.G. Lima were recipients of fellowships from FAPESP.
 Address for correspondence: A.M.L. Castrucci, Departamento de Fisiologia, Instituto de Biociências, USP, Rua do Matão, Travessa 14, 101, Sala 317, 05508-900 São Paulo, SP, Brasil. Fax: +55-11-3091-7422. E-mail: amdlcast@ib.usp.br
Address for correspondence: A.M.L. Castrucci, Departamento de Fisiologia, Instituto de Biociências, USP, Rua do Matão, Travessa 14, 101, Sala 317, 05508-900 São Paulo, SP, Brasil. Fax: +55-11-3091-7422. E-mail: amdlcast@ib.usp.br
Received April 11, 2010. Accepted August 11, 2010. Available online August 27, 2010. Published September 13, 2010.
The Brazilian Journal of Medical and Biological Research is partially financed by
- 1. Yanagisawa M, Kurihara H, Kimura S, Tomobe Y, Kobayashi M, Mitsui Y, et al. A novel potent vasoconstrictor peptide produced by vascular endothelial cells. Nature 1988; 332: 411-415.
- 2. Ducancel F. The sarafotoxins. Toxicon 2002; 40: 1541-1545.
- 3. Kochva E, Viljoen CC, Botes DP. A new type of toxin in the venom of snakes of the genus Atractaspis (Atractaspidinae). Toxicon 1982; 20: 581-592.
- 4. Yohn JJ, Morelli JG, Walchak SJ, Rundell KB, Norris DA, Zamora MR. Cultured human keratinocytes synthesize and secrete endothelin-1. J Invest Dermatol 1993; 100: 23-26.
- 5. Hirobe T. Role of keratinocyte-derived factors involved in regulating the proliferation and differentiation of mammalian epidermal melanocytes. Pigment Cell Res 2005; 18: 2-12.
- 6. Yada Y, Higuchi K, Imokawa G. Effects of endothelins on signal transduction and proliferation in human melanocytes. J Biol Chem 1991; 266: 18352-18357.
- 7. Imokawa G, Miyagishi M, Yada Y. Endothelin-1 as a new melanogen: coordinated expression of its gene and the tyrosinase gene in UVB-exposed human epidermis. J Invest Dermatol 1995; 105: 32-37.
- 8. Hayashi H, Nakamura S, Fujii R. The endothelin receptors that mediate aggregation of pigment in fish melanophores. Comp Biochem Physiol B 1996; 115: 143-152.
- 9. Murata N, Fujii R. Pigment-aggregating action of endothelins on medaka xantophores. Zool Sci 2000; 17: 853-862.
- 10. Arai H, Hori S, Aramori I, Ohkubo H, Nakanishi S. Cloning and expression of a cDNA encoding an endothelin receptor. Nature 1990; 348: 730-732.
- 11. Sakurai T, Yanagisawa M, Takuwa Y, Miyazaki H, Kimura S, Goto K, et al. Cloning of a cDNA encoding a non-isopeptide-selective subtype of the endothelin receptor. Nature 1990; 348: 732-735.
- 12. Karne S, Jayawickreme CK, Lerner MR. Cloning and characterization of an endothelin-3 specific receptor (ETC receptor) from Xenopus laevis dermal melanophores. J Biol Chem 1993; 268: 19126-19133.
- 13. Filadelfi AM, Ramanzini GC, Visconti MA, Castrucci AM. The endothelin/sarafotoxin-induced increase of the proliferation of undifferentiated and DMSO-differentiated GEM-81 goldfish erythrophoroma cells is mediated by ETB receptors. Pigment Cell Res 2004; 17: 480-487.
- 14. Scarparo AC, Isoldi MC, de Lima LH, Visconti MA, Castrucci AM. Expression of endothelin receptors in frog, chicken, mouse and human pigment cells. Comp Biochem Physiol A Mol Integr Physiol 2007; 147: 640-646.
- 15. Ambar I, Sokolovsky M. Endothelin receptors stimulate both phospholipase C and phospholipase D activities in different cell lines. Eur J Pharmacol 1993; 245: 31-41.
- 16. Kasuya Y, Abe Y, Hama H, Sakurai T, Asada S, Masaki T, et al. Endothelin-1 activates mitogen-activated protein kinases through two independent signalling pathways in rat astrocytes. Biochem Biophys Res Commun 1994; 204: 1325-1333.
- 17. Fujii R. The regulation of motile activity in fish chromatophores. Pigment Cell Res 2000; 13: 300-319.
- 18. Ban E, Kasai A, Sato M, Yokozeki A, Hisatomi O, Oshima N. The signaling pathway in photoresponses that may be mediated by visual pigments in erythrophores of Nile tilapia. Pigment Cell Res 2005; 18: 360-369.
- 19. Im LH, Isoldi MC, Scarparo AC, Visconti MA, Castrucci AM. Rhythmic expression, light entrainment and alpha-MSH modulation of rhodopsin mRNA in a teleost pigment cell line. Comp Biochem Physiol A Mol Integr Physiol 2007; 147: 691-696.
- 20. Miyashita Y, Moriya T, Kubota T, Yamada K, Asami K. Expression of opsin molecule in cultured murine melanocyte. J Investig Dermatol Symp Proc 2001; 6: 54-57.
- 21. Chen S, Zack DJ. Ret 4, a positive acting rhodopsin regulatory element identified using a bovine retina in vitro transcription system. J Biol Chem 1996; 271: 28549-28557.
- 22. Zack DJ, Bennett J, Wang Y, Davenport C, Klaunberg B, Gearhart J, et al. Unusual topography of bovine rhodopsin promoter-lacZ fusion gene expression in transgenic mouse retinas. Neuron 1991; 6: 187-199.
- 23. Nie Z, Chen S, Kumar R, Zack DJ. RER, an evolutionarily conserved sequence upstream of the rhodopsin gene, has enhancer activity. J Biol Chem 1996; 271: 2667-2675.
- 24. Fitzgibbon J, Hope A, Slobodyanyuk SJ, Bellingham J, Bowmaker JK, Hunt DM. The rhodopsin-encoding gene of bony fish lacks introns. Gene 1995; 164: 273-277.
- 25. Kawamura S, Takeshita K, Tsujimura T, Kasagi S, Matsumoto Y. Evolutionarily conserved and divergent regulatory sequences in the fish rod opsin promoter. Comp Biochem Physiol B Biochem Mol Biol 2005; 141: 391-399.
- 26. Liu Y, Shen Y, Rest JS, Raymond PA, Zack DJ. Isolation and characterization of a zebrafish homologue of the cone rod homeobox gene. Invest Ophthalmol Vis Sci 2001; 42: 481-487.
- 27. Bremnes T, Paasche JD, Mehlum A, Sandberg C, Bremnes B, Attramadal H. Regulation and intracellular trafficking pathways of the endothelin receptors. J Biol Chem 2000; 275: 17596-17604.
- 28. Cheng TH, Shih NL, Chen CH, Lin H, Liu JC, Chao HH, et al. Role of mitogen-activated protein kinase pathway in reactive oxygen species-mediated endothelin-1-induced beta-myosin heavy chain gene expression and cardiomyocyte hypertrophy. J Biomed Sci 2005; 12: 123-133.
- 29. Rodriguez-Vita J, Ruiz-Ortega M, Ruperez M, Esteban V, Sanchez-Lopez E, Plaza JJ, et al. Endothelin-1, via ETA receptor and independently of transforming growth factor-beta, increases the connective tissue growth factor in vascular smooth muscle cells. Circ Res 2005; 97: 125-134.
- 30. Muldoon LL, Rodland KD, Forsythe ML, Magun BE. Stimulation of phosphatidylinositol hydrolysis, diacylglycerol release, and gene expression in response to endothelin, a potent new agonist for fibroblasts and smooth muscle cells. J Biol Chem 1989; 264: 8529-8536.
- 31. Komuro I, Kurihara H, Sugiyama T, Yoshizumi M, Takaku F, Yazaki Y. Endothelin stimulates c-fos and c-myc expression and proliferation of vascular smooth muscle cells. FEBS Lett 1988; 238: 249-252.
- 32. Pribnow D, Muldoon LL, Fajardo M, Theodor L, Chen LY, Magun BE. Endothelin induces transcription of fos/jun family genes: a prominent role for calcium ion. Mol Endocrinol 1992; 6: 1003-1012.
- 33. Bobik A, Grooms A, Millar JA, Mitchell A, Grinpukel S. Growth factor activity of endothelin on vascular smooth muscle. Am J Physiol 1990; 258: C408-C415.
- 34. Koide M, Kawahara Y, Tsuda T, Ishida Y, Shii K, Yokoyama M. Endothelin-1 stimulates tyrosine phosphorylation and the activities of two mitogen-activated protein kinases in cultured vascular smooth muscle cells. J Hypertens 1992; 10: 1173-1182.
- 35. Stork PJ, Schmitt JM. Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol 2002; 12: 258-266.
- 36. Kolch W. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem J 2000; 351 (Part 2): 289-305.
- 37. Naicker S, Bhoola KD. Endothelins: vasoactive modulators of renal function in health and disease. Pharmacol Ther 2001; 90: 61-88.
- 38. Imokawa G, Yada Y, Kimura M. Signalling mechanisms of endothelin-induced mitogenesis and melanogenesis in human melanocytes. Biochem J 1996; 314 (Part 1): 305-312.
- 39. Palumbo AP, Rossino P, Comoglio PM. Bombesin stimulation of c-fos and c-myc gene expression in cultures of Swiss 3T3 cells. Exp Cell Res 1986; 167: 276-280.
Correspondence and Footnotes
Publication Dates
-
Publication in this collection
03 Sept 2010 -
Date of issue
Sept 2010
History
-
Accepted
11 Aug 2010 -
Received
11 Apr 2010