Abstract
The actions of thyroid hormone (TH) on pancreatic beta cells have not been thoroughly explored, with current knowledge being limited to the modulation of insulin secretion in response to glucose, and beta cell viability by regulation of pro-mitotic and pro-apoptotic factors. Therefore, the effects of TH on proinsulin gene expression are not known. This led us to measure: a) proinsulin mRNA expression, b) proinsulin transcripts and eEF1A protein binding to the actin cytoskeleton, c) actin cytoskeleton arrangement, and d) proinsulin mRNA poly(A) tail length modulation in INS-1E cells cultured in different media containing: i) normal fetal bovine serum - FBS (control); ii) normal FBS plus 1 µM or 10 nM T3, for 12 h, and iii) FBS depleted of TH for 24 h (Tx). A decrease in proinsulin mRNA content and attachment to the cytoskeleton were observed in hypothyroid (Tx) beta cells. The amount of eEF1A protein anchored to the cytoskeleton was also reduced in hypothyroidism, and it is worth mentioning that eEF1A is essential to attach transcripts to the cytoskeleton, which might modulate their stability and rate of translation. Proinsulin poly(A) tail length and cytoskeleton arrangement remained unchanged in hypothyroidism. T3 treatment of control cells for 12 h did not induce any changes in the parameters studied. The data indicate that TH is important for proinsulin mRNA expression and translation, since its total amount and attachment to the cytoskeleton are decreased in hypothyroid beta cells, providing evidence that effects of TH on carbohydrate metabolism also include the control of proinsulin gene expression.
Proinsulin mRNA; eEF1A protein; Cytoskeleton; Hypothyroidism; INS-1E cells
Abstract
Introduction
Material and Methods
Results
Discussion
References
Acknowledgments
Braz J Med Biol Res, October 2011, Volume 44(10) 1060-1067
Hypothyroidism decreases proinsulin gene expression and the attachment of its mRNA and eEF1A protein to the actin cytoskeleton of INS-1E cells
F. Goulart-Silva, C. Serrano-Nascimento and  Correspondence and Footnotes
Correspondence and Footnotes
 M.T. Nunes
M.T. Nunes
Departamento de Fisiologia e Biofísica, Instituto de Ciências Biomédicas, Universidade de São Paulo, São Paulo, SP, Brasil
 Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
The actions of thyroid hormone (TH) on pancreatic beta cells have not been thoroughly explored, with current knowledge being limited to the modulation of insulin secretion in response to glucose, and beta cell viability by regulation of pro-mitotic and pro-apoptotic factors. Therefore, the effects of TH on proinsulin gene expression are not known. This led us to measure: a) proinsulin mRNA expression, b) proinsulin transcripts and eEF1A protein binding to the actin cytoskeleton, c) actin cytoskeleton arrangement, and d) proinsulin mRNA poly(A) tail length modulation in INS-1E cells cultured in different media containing: i) normal fetal bovine serum - FBS (control); ii) normal FBS plus 1 µM or 10 nM T3, for 12 h, and iii) FBS depleted of TH for 24 h (Tx). A decrease in proinsulin mRNA content and attachment to the cytoskeleton was observed in hypothyroid (Tx) beta cells. The amount of eEF1A protein anchored to the cytoskeleton was also reduced in hypothyroidism, and it is worth mentioning that eEF1A is essential to attach transcripts to the cytoskeleton, which might modulate their stability and rate of translation. Proinsulin poly(A) tail length and cytoskeleton arrangement remained unchanged in hypothyroidism. T3 treatment of control cells for 12 h did not induce any changes in the parameters studied. The data indicate that TH is important for proinsulin mRNA expression and translation, since its total amount and attachment to the cytoskeleton are decreased in hypothyroid beta cells, providing evidence that effects of TH on carbohydrate metabolism also include the control of proinsulin gene expression.
Key words: Proinsulin mRNA; eEF1A protein; Cytoskeleton; Hypothyroidism; INS-1E cells
Thyroid hormone (TH) is known to modulate developmental and metabolic processes by interacting with specific binding sites located in the nucleus, cytoplasm, organelles, and plasma membrane (1-5). It is well known that TH affects the metabolic rate by controlling lipid, protein and carbohydrate synthesis and degradation (6,7), actions that are also shared by insulin and growth hormone (GH) (8-11). Besides, TH is known to induce GH gene expression, which might indirectly account for its metabolic effects.
On the other hand, the action of TH on insulin gene expression remains uncertain, as the available data are scarce and sometimes conflicting. Indeed, one study has shown that T3 enhances proinsulin mRNA levels during pancreatic beta cell differentiation from the human duct cell line (hPANC-1), whereas other studies have shown the opposite effects, that is, T3 and T4 leading to a decrease in proinsulin gene expression in RIN-m5F cells and rat pancreatic beta cells, respectively (12-14).
The regulation of insulin gene expression is an important step in carbohydrate metabolism, and even though several hormones have been shown to modulate it (15-17), the participation of TH in this process has been poorly explored. In fact, hyperthyroid and hypothyroid states have been associated with the impairment of glucose homeostasis, which might alter insulin synthesis and release, blunting a possible direct effect of TH on the proinsulin gene expression. Moreover, recent evidence that TH might post-transcriptionally regulate the expression of certain genes indicates that it can rapidly interfere with the stability and translation rate of distinct mRNAs (18,19). This could be another mechanism by which TH could affect proinsulin gene expression, which has never been investigated.
In fact, the post-transcriptional control of gene expression by TH includes effects on mRNA polyadenylation, which could be enhanced or decreased, increasing or decreasing transcript stability and translation rate, as shown for GH mRNA and TSH mRNA, respectively (18,19). In addition, it is known that the poly(A) tail of the transcripts plays an important role in their attachment to the cytoskeleton, which would also help to protect them from RNases, leading to repercussion on their half-lives (20).
The transcript attachment to the cytoskeleton is mediated by specific proteins, which recognize binding sites in mRNA molecules and simultaneously in proteins of the cytoskeleton. Despite the fact that several proteins have already been identified as mediators of this binding process, the eukaryotic elongation factor 1 A (eEF1A), a primary protein involved in protein synthesis, has been shown by Liu et al. (21) to be able to attach transcripts to the cytoskeleton.
Therefore, the present study attempted to investigate in INS-1E cells the effects of TH on proinsulin gene expression, and its possible post-transcriptional effects on: a) proinsulin transcript poly(A) tail length and attachment to the cytoskeleton, b) eEF1A protein binding to the cytoskeleton, and c) cytoskeleton organization. Taken together, these data should provide insight into the control of proinsulin gene expression by TH, and strengthen the existing evidence for the metabolic actions of TH.
Material
Triiodo-L-thyronine (T3), Triton X-100, L-glutamine, and anti-ß-actin antibody were purchased from Sigma Chemical Co. (USA). The anti-α-actinin antibody was purchased from Santa Cruz Biotechnology, Inc. (USA), and anti-EF1A was purchased from Upstate Biotechnology Inc. (USA). Phenol, guanidine isothiocyanate, sodium N-lauryl sarcosine, diethyl pyrocarbonate (DEPC), agarose, N-morpholino-propanesulfonic acid buffer (MOPS), sodium dodecyl sulfate, ethidium bromide, BSA, oligo(dT) and dNTP, RNase-free DNase I, DNA ladder, HEPES, and ß-mercaptoethanol were purchased from Invitrogen Life Technologies, Inc. (USA). The anti-mouse and anti-rabbit horseradish peroxidase antibodies were purchased from Jackson Laboratories (USA). Rhodamine-phalloidin was purchased from Molecular Probes (USA). Reverse transcriptase (M-MLV), 5X First-Strand buffer, GoTaq® DNA Polymerase and 5X Green GoTaq® Flexi Buffer were purchased from Promega (USA). SYBR® Green PCR master mix was purchased from Applied Biosystems (USA). The enhanced chemiluminescence (ECL) kit, protein molecular weight markers and the Bio-Rad protein assay kit were purchased from Bio-Rad Laboratórios Brasil Ltda. (Brazil). Heat-inactivated fetal bovine serum (FBS), phosphate-buffered saline, trypsin/EDTA solution, RPMI 1640 culture medium, and sodium pyruvate were purchased from Vitrocell Comércio de Produtos para Laboratórios Ltda. (Brazil). Tissue culture materials were purchased from TPP Techno Plastic Products AG (Switzerland). All other reagents were purchased from Labsynth (Brazil).
Cell culture
The rat INS-1E insulinoma cell line was kindly provided by Prof. Ângelo Rafael Carpinelli (Universidade de São Paulo, São Paulo, SP, Brazil) and cultured in RPMI 1640 with 11 mM glucose, supplemented with 10% FBS, 10 mM HEPES, 1 mM sodium pyruvate, 2 mM L-glutamine, 50 U/mL penicillin, 50 µg/mL streptomycin, and 50 µm 2-mercaptoethanol. Cells were cultured between passages 63-72 and were grown at 37°C in 95% humidified air with 5% CO2. Cells were used for experiments at 80-90% confluence. To perform the experiments, the cells were maintained in a) normal FBS (control group), b) normal FBS plus 1 µM or 10 nM T3, for 12 h, and c) FBS depleted from TH, for 24 h (Tx group). The following parameters were then evaluated: a) proinsulin gene expression by real-time PCR, b) proinsulin transcripts and eEF1A protein attached to the actin cytoskeleton, c) actin cytoskeleton rearrangement by actin fractionation, and d) proinsulin mRNA poly(A) tail length by rapid amplification of cDNA ends - poly(A) test (RACE-PAT). TH depletion from FBS was carried out according to Hollenberg et al. (22).
Cell viability assay
INS-1E cells (1 x 106 cells/mL) were resuspended in 300 µL PBS and 30 µL propidium iodide solution (20 µg/mL in PBS) was added. The percentage of viable cells in each sample was determined using a FACSCalibur flow cytometer (Becton-Dickinson, USA). The flow cytometer was frequently checked, calibrated and tested for quality control. Propidium iodide is a highly water-soluble fluorescent compound that cannot pass through intact membranes, and is generally excluded from viable cells. It binds to DNA by intercalating between the bases with little or no sequence preference. Fluorescence was measured using the FL2 channel (Orange-red fluorescence-585/42 nm). Ten thousand events were analyzed per experiment. Cells with propidium iodide fluorescence were then evaluated using the Cell Quest software (Becton-Dickinson) (23).
Measurement of proinsulin gene expression by real-time PCR
Total RNA was extracted by the guanidine-phenol-chloroform method described by Chomczynski and Sacchi (24) and quantified by spectrophotometry. Samples were treated with RNase-free DNase I (Invitrogen), and 1 µg RNA was reverse-transcribed in a reaction containing oligo(dT) (100 µg/mL), 10 mM dNTP, 5X First-Strand buffer, and 2 µL 200 U M-MLV reverse transcriptase (Invitrogen). For real-time PCR amplification, 2 µL of each reverse transcription (RT) product was diluted in 200 µL DEPC water, and 1 µL was added to a reaction buffer containing 5 µL SYBR® Green PCR master mix (Applied Biosystems) and 900 nM primers, in a final volume of 12.5 µL per sample. The sequences of the primers used are listed in Table 1 and were designed using the Primer-BLAST tool. The reaction conditions consisted of two steps at 50°C for 2 min and at 95°C for 2 min, followed by 45 cycles of three steps: 15 s denaturation at 95°C, 60 s annealing at 60°C, and 15 s at 72°C. The threshold cycle (CT) of the real-time PCR was measured in duplicate for each sample. The CT value represents the PCR cycle number at which the increasing fluorescence intensity of amplified RT-PCR products of the target gene is exponential, and is inversely correlated with the amount of the target mRNA in the sample. A DCT value was calculated for each sample by subtracting the CT value of the reference gene from the CT value of the gene of interest. To reduce interassay variation, all samples were normalized to the DCT value of a control sample, the calibrator one, derived from the same control, for all assays (DDCT). The relative expression of mRNAs was calculated using the 2-DDCT method, and is reported as arbitrary units (AU). The reference gene used for real-time PCR was RPL19, which presented the most uniform expression in response to different treatments.
Measurement of proinsulin mRNA poly(A) tail length by RACE-PAT
Proinsulin mRNA poly(A) tail length was measured by the RACE-PAT according to a standard protocol (25,26). Briefly, 200 ŋg of an oligo(dT) anchor (5'-GCGAGCTCCGCGGCC GCG-T12) was added to 2 µg total RNA in a sterile RNase-free microfuge tube. The samples were denatured at 65°C for 5 min, transferred to 42°C and reverse-transcribed for 60 min using a mix containing 10 mM of each dNTP, 5X First-Strand buffer, 10 mM DTT, 1 µL reverse transcriptase M-MLV (Invitrogen). Inactivation was performed at 70°C for 15 min. Three microliters of the RT reaction product was mixed with a reaction buffer containing 25 mM MgCl2, 10 mM of each dNTP, 25 pmol of each primer sense (Table 1), 1.25 U of GoTaq® DNA Polymerase and 5X Green GoTaq® Flexi Buffer (Promega). The reaction was performed for 40 cycles, each consisting of 95°C for 30 s (denaturation), 64°C for 1 min (annealing), and 72°C for 1 min (extension), followed by a final 7-min elongation step at 72°C. The PCR-amplified products were analyzed by denaturing 2.5% agarose gel electrophoresis and ethidium bromide staining. Amplicon sizes were estimated by densitometry and compared with a 100-bp DNA ladder (Invitrogen Life Technologies) using the ImageQuant TL software (Version 2005 Amersham Biosciences, USA). The top of the smear indicates the longest amplified fragment, which represents the poly(A) tail size plus 405 bases upstream, according to the proinsulin primer used. The changes in poly(A) tail length were detected by analyzing the length of cDNA smears obtained in all experimental groups studied.
Measurement of polymerized (F) and non-polymerized (G) actin content by Western blotting analysis
Actin fractionation. The protocol was adapted from Posern et al. (27), Haller et al. (28), who included phalloidin in the lysis buffer described by Posern, and Liu et al. (21), who evaluated eEF1A binding to the actin cytoskeleton. Cells were lysed in 0.75 mL lysis buffer containing 50 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 20 mM HEPES, pH 7.9, and 132 nM phalloidin, a fungal toxin that specifically binds to polymerized actin (F-actin) and protects it from depolymerization. After 10 min, the homogenate was centrifuged at 100,000 g for 30 min at 4°C, and the supernatant (non-polymerized actin) and pellet fractions (polymerized actin) were obtained. The pellet, which contains the F-actin fraction, was resuspended in 0.2 mL actin lysis buffer and then sonicated. The amount of proteins in the pellet and supernatant was determined by the method of Bradford (29).
Measurement of proinsulin transcripts and eEF1A binding to F-actin
In order to determine the amount of eEF1A and proinsulin mRNA bound to F-actin, the INS-1E cells were lysed in 0.75 mL lysis buffer containing 50 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 20 mM HEPES, pH 7.9, and 132 nM phalloidin. G- and F-actin were separated by ultracentrifugation at 100,000 g for 30 min at 4°C. Total RNA was extracted from the F-actin fraction by the guanidine-phenol-chloroform method and submitted to real-time PCR. eEF1A protein was also measured in the F-actin fraction by immunoblotting (18).
Immunoblotting analysis
Thirty micrograms of protein from each sample was treated with Laemmli buffer (30), submitted to 10% SDS-PAGE, and transferred to a nitrocellulose membrane by electroblotting for 60 min at 100 V. Nonspecific protein binding to the membrane was reduced by preincubation with blocking buffer (5% nonfat dry milk, 2.7 mM KCl, 137 mM NaCl, 8 mM NaHPO4, 1.4 mM KPO4, and 0.1% Tween 20) overnight at 4°C. The membrane was incubated with the monoclonal anti-ß-actin (1:5000) or anti-eEF1A (1:1000) antibody in blocking buffer, for 2 h at room temperature, followed by incubation with the appropriate secondary peroxidase-conjugated antibody (1:5000) in 2.7 mM KCl, 137 mM NaCl, 8 mM NaHPO4, 1.4 mM KPO4, and 0.1% Tween 20 buffer, for 90 min at room temperature. After washing the membrane, bands were detected using the ECL kit. Blots were analyzed by scanning densitometry and quantified using the Image Master-1D-Pharmacia Biotech SW software (Pharmacia Biotech, Sweden). Membranes were also incubated with monoclonal anti-α-actinin antibody (1:1000), which was used as an internal control, under the same conditions as described above. The results are reported as AU normalized by α-actinin protein.
Statistical analysis
Data are reported as means ± SEM and were submitted to one-way analysis of variance (ANOVA) followed by the Student-Newman-Keuls post-test. Differences were considered to be significant when P < 0.05.
Thyroid hormone did not alter INS-1E cell viability
The analysis of membrane integrity of INS-1E cells showed that the absence of TH or T3 treatment did not alter cell viability, since the percentage of control cells (89.62 ± 2.05), hypothyroid (94.57 ± 0.67) or control cells treated with 1 µM T3 (95.96 ± 0.37), or 10 nM T3 (97.82 ± 1.96) with an intact plasma membrane did not change.
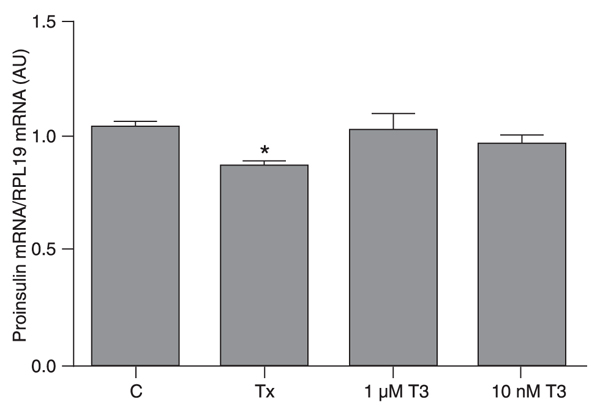
Proinsulin gene expression is decreased in hypothyroid pancreatic beta cells
The beta cells maintained in the hypothyroid condition had lower proinsulin mRNA levels compared to the control group. In contrast, control cells treated with T3 for 12 h did not show an alteration in proinsulin mRNA expression, which remained similar to those observed in the control group (Figure 1).
Hypothyroidism decreases proinsulin mRNAs bound to the actin cytoskeleton
To determine proinsulin mRNA association with the cytoskeleton, G-actin and F-actin were separated by ultracentrifugation as described above. In the absence of TH, beta cells showed less proinsulin mRNA bound to the actin cytoskeleton; however, control cells incubated with T3 did not show an alteration of the amount of proinsulin mRNA anchored to the cytoskeleton (Figure 2).
Hypothyroidism decreases the attachment rate of eEF1A to the actin cytoskeleton
eEF1A was also measured in the F-actin fraction after cytoskeleton fractionation. When pancreatic beta cells were maintained in the hypothyroid condition, there was a reduction of the eEF1A amount attached to the actin cytoskeleton when compared to the control group.
In contrast, when the cells were incubated with T3 for 12 h no significant change was observed in the amount of eEF1A attached to the actin cytoskeleton (Figure 3).
The cytoskeleton remains unchanged after thyroid hormone treatment
G-actin and F-actin from the cytoskeleton were measured in the experimental and control groups. The amounts of the proteins were unchanged after TH withdrawal or T3 addition. Data for F-actin (pellet fraction) are presented in Figure 4A and for G-actin (supernatant fraction) in Figure 4B.
Proinsulin poly(A) tail length remains unchanged after thyroid hormone treatment
The data obtained from RACE-PAT are shown in Figure 5. The poly(A) tail length of proinsulin transcripts from pancreatic beta cells did not change after TH withdrawal or T3 addition, being similar for the experimental and control groups.
Hypothyroidism decreases proinsulin gene expression. INS-1E cells were divided into control (C), hypothyroid (Tx) and control plus T3 (1 µM or 10 nM T3) groups. Proinsulin gene expression was measured by real-time PCR and the values obtained were corrected by RPL19. Data are reported as means ± SEM for three independent experiments (N = 4 per experiment), expressed in arbitrary units (AU). *P < 0.05 vs all other groups (one-way ANOVA followed by the Student-Newman-Keuls post-test).
Hypothyroidism decreases proinsulin transcripts anchored to the actin cytoskeleton. INS-1E cells were divided into control (C), hypothyroid (Tx) and control plus T3 (1 µM or 10 nM T3) groups. F-actin from the cytoskeleton was preserved and separated by ultracentrifugation. Total RNA was then extracted from the F-actin network, and proinsulin gene expression was measured by real-time PCR being normalized by RPL19 expression. Data are reported as means ± SEM for two independent experiments (N = 6 per experiment), expressed in arbitrary units (AU). *P < 0.05 vs all other groups (one-way ANOVA followed by the Student-Newman-Keuls post-test).
Hypothyroidism decreases the amount of eEF1A protein anchored to the cytoskeleton. INS-1E cells were divided into control (C), hypothyroid (Tx) and control plus T3 (1 µM or 10 nM T3) groups. F-actin from the cytoskeleton was preserved and separated by ultracentrifugation. Proteins were then extracted from F-actin and submitted to electrophoresis and Western blotting analysis for eEF1A, with the values obtained being normalized by α-actinin. Blots of representative experiments are shown at the top of the figure. Data are reported as means ± SEM for two independent experiments (N = 6 per experiment), expressed in arbitrary units (AU). *P < 0.05 vs all other groups (one-way ANOVA followed by the Student-Newman-Keuls post-test).
Thyroid hormone does not affect cytoskeleton rearrangement. Panels A and B show F-actin (pellet fraction) and G-actin (supernatant fraction), respectively. INS-1E cells were divided into control (C), hypothyroid (Tx) and control plus T3 (1 µM or 10 nM T3) groups. Total actin protein content was determined by Western blotting and the values obtained were normalized by α-actinin. Blots of representative experiments are shown at the top of Figures A and B. Data are reported as means ± SEM for four independent experiments (N = 4 per experiment), expressed in arbitrary units (AU). No significant differences were observed between groups.
Thyroid hormone does not affect proinsulin transcript poly(A) tail length. The proinsulin mRNA poly(A) tail length was determined by rapid amplification of cDNA ends for the poly(A) test (RACE-PAT) in INS-1E cells divided into control (C), hypothyroid (Tx) and control plus T3 (1 µM or 10 nM T3) groups. The smearing pattern of ethidium bromide-stained PCR products obtained by RACE-PAT is shown at the top of the figure. The horizontal line delimits the heaviest PCR fragments generated in the samples. The maximal size of the amplicons, corresponding to the top of the smear of each sample (in bp), is shown at the top. Data are reported as means ± SEM for two independent experiments (N = 3 per experiment), expressed in bp. No significant differences were observed between groups. RACE-PAT = rapid amplification of cDNA ends - poly(A) test.
The present study shows that TH modulates proinsulin gene expression, since INS-1E cells cultured in its absence presented a decrease in proinsulin gene expression, as shown by the reduced amount of its transcripts. However, T3 addition to pancreatic beta cells in culture did not modify the amount of proinsulin mRNA. This differential modulation indicates that the presence of TH at physiological concentrations is essential for the establishment of some biological processes, as demonstrated by proinsulin mRNA expression. Indeed, these data agree with reports that showed an increase of myostatin gene expression in hypothyroid rats, even though no changes in myostatin mRNA levels were detected in L-T4-treated rats (31). However, the mechanisms underlying these alterations, observed only in the hypothyroid condition, are unknown.
RNAs are synthesized in the nucleus and exported to the cytoplasm, being addressed to specific regions inside the cells through cytoskeleton proteins (32,33). Transcripts are then attached to the cytoskeleton, which modulates their turnover rate and, consequently, the total amount of mRNA. Besides the alteration in proinsulin gene expression, the amount of its transcripts attached to the cytoskeleton is decreased in hypothyroidism, which might interfere with proinsulin mRNA stability, since the anchoring of mRNAs to the cytoskeleton increases or decreases transcript stability by a mechanism that most likely involves the regulation of the activity of specific RNases (34,35).
The interaction between transcripts and the cytoskeleton is a process mediated by proteins that bind to mRNAs, as well as to cytoskeleton proteins at different sites, at the same time. Several proteins are thought to possess this ability. In the present study, we evaluated the amount of eEF1A attached to the cytoskeleton, since studies from our laboratory have shown that hypothyroidism decreases the association between eEF1A and actin in rat pituitary, an event promptly reversed after acute T3 administration (18).
Indeed, as shown in pituitaries from hypothyroid rats, the amount of eEF1A anchored to the cytoskeleton is reduced in pancreatic beta cells, suggesting that eEF1A may intermediate the anchoring of proinsulin transcripts to the actin cytoskeleton. It should be noted that the alterations in proinsulin mRNA and eEF1A levels attached to the cytoskeleton are restricted to beta cells maintained in the hypothyroid condition, as these parameters were not changed after T3 treatment, reinforcing the importance of the euthyroid condition for the maintenance of normal proinsulin gene expression and its turnover. The fact that the attachment of both eEF1A and proinsulin mRNA to actin is decreased in the hypothyroidism condition does not necessarily indicate that these processes are directly linked, even though each one, per se, leads to the reduction of transcript stability, as described (18,21), and points to a post-transcriptional action of TH on the control of proinsulin gene expression.
Despite being the main structure for anchoring proinsulin transcripts and eEF1A, the cytoskeleton of the pancreatic beta cell is not altered after TH treatment since the amounts of G- and F-actin remain similar in the hypothyroid and hyperthyroid conditions. Hence, the decreased proinsulin mRNA and eEF1A content in the cytoskeleton fraction may indicate that hypothyroidism impairs the attachment of these molecules to the cytoskeleton, but it appears that these events are not associated with cytoskeleton disarrangement.
Moreover, the cytoskeleton may be capable of intermediating the approach of transcripts to poly(A) polymerase, a process that leads to an increase in transcript poly(A) tail length by insertion of adenine residues. This process increases mRNA stability, as degradation of transcripts by RNases diminishes, enhancing the translation rate and protein accumulation (36).
Despite the regulation of several transcript poly(A) tail length by TH, in the present study, the number of adenine residues in the proinsulin mRNA tail remained unaltered in cells cultured in hypothyroid medium as well as after T3 treatment, which indicates that TH regulates proinsulin mRNA stability probably by anchoring the transcripts to the cytoskeleton, without involvement of the poly(A) tail.
The data presented here show that at least part of the modulation of proinsulin gene expression by TH seems to occur at the post-transcriptional level, since proinsulin mRNA turnover depends on the attachment of proinsulin transcripts and eEF1A to the cytoskeleton, which is a biological process that occurs after the gene transcription, interfering with insulin production by the cells. However, we cannot rule out the possibility that TH might act physiologically inducing proinsulin gene transcription. These data should be considered when the action of TH on carbohydrate metabolism is under investigation.
1. Bergh JJ, Lin HY, Lansing L, Mohamed SN, Davis FB, Mousa S, et al. Integrin alphaVbeta3 contains a cell surface receptor site for thyroid hormone that is linked to activation of mitogen-activated protein kinase and induction of angiogenesis. Endocrinology 2005; 146: 2864-2871.
2. Cheng SY, Gong QH, Parkison C, Robinson EA, Appella E, Merlino GT, et al. The nucleotide sequence of a human cellular thyroid hormone binding protein present in endoplasmic reticulum. J Biol Chem 1987; 262: 11221-11227.
3. Psarra AM, Solakidi S, Sekeris CE. The mitochondrion as a primary site of action of steroid and thyroid hormones: presence and action of steroid and thyroid hormone receptors in mitochondria of animal cells. Mol Cell Endocrinol 2006; 246: 21-33.
4. Yen PM. Physiological and molecular basis of thyroid hormone action. Physiol Rev 2001; 81: 1097-1142.
5. Bhargava M, Lei J, Mariash CN, Ingbar DH. Thyroid hormone rapidly stimulates alveolar Na,K-ATPase by activation of phosphatidylinositol 3-kinase. Curr Opin Endocrinol Diabetes Obes 2007; 14: 416-420.
6. Kim B. Thyroid hormone as a determinant of energy expenditure and the basal metabolic rate. Thyroid 2008; 18: 141-144.
7. Kaminsky P, Klein M, Duc M. [Control of muscular bioenergetics by the thyroid hormones]. Presse Med 1993; 22: 774-778.
8. Lukens FD. Insulin and protein metabolism. Diabetes 1964; 13: 451-461.
9. Moller N, Jorgensen JO. Effects of growth hormone on glucose, lipid, and protein metabolism in human subjects. Endocr Rev 2009; 30: 152-177.
10. Foufelle F, Hegarty B, Bobard A, Hainault I, Bossard P, Ferre P. [A new role for insulin in the control of energetic metabolism in liver]. Med Sci 2005; 21: 569-571.
11. Vijayakumar A, Novosyadlyy R, Wu Y, Yakar S, LeRoith D. Biological effects of growth hormone on carbohydrate and lipid metabolism. Growth Horm IGF Res 2010; 20: 1-7.
12. Misiti S, Anastasi E, Sciacchitano S, Verga FC, Panacchia L, Bucci B, et al. 3,5,3'-Triiodo-L-thyronine enhances the differentiation of a human pancreatic duct cell line (hPANC-1) towards a beta-cell-like phenotype. J Cell Physiol 2005; 204: 286-296.
13. Fernandez-Mejia C, Davidson MB. Regulation of glucokinase and proinsulin gene expression and insulin secretion in RIN-m5F cells by dexamethasone, retinoic acid, and thyroid hormone. Endocrinology 1992; 130: 1660-1668.
14. Fukuchi M, Shimabukuro M, Shimajiri Y, Oshiro Y, Higa M, Akamine H, et al. Evidence for a deficient pancreatic beta-cell response in a rat model of hyperthyroidism. Life Sci 2002; 71: 1059-1070.
15. Yildiz BO, Haznedaroglu IC. Rethinking leptin and insulin action: therapeutic opportunities for diabetes. Int J Biochem Cell Biol 2006; 38: 820-830.
16. Holst JJ. The physiology of glucagon-like peptide 1. Physiol Rev 2007; 87: 1409-1439.
17. Strowski MZ, Parmar RM, Blake AD, Schaeffer JM. Somatostatin inhibits insulin and glucagon secretion via two receptor subtypes: an in vitro study of pancreatic islets from somatostatin receptor 2 knockout mice. Endocrinology 2000; 141: 111-117.
18. Silva FG, Giannocco G, Luchessi AD, Curi R, Nunes MT. T3 acutely increases GH mRNA translation rate and GH secretion in hypothyroid rats. Mol Cell Endocrinol 2010; 317: 1-7.
19. Goulart-Silva F, de Souza PB, Nunes MT. T3 rapidly modulates TSHbeta mRNA stability and translational rate in the pituitary of hypothyroid rats. Mol Cell Endocrinol 2011; 332: 277-282.
20. Bonneau AM, Darveau A, Sonenberg N. Effect of viral infection on host protein synthesis and mRNA association with the cytoplasmic cytoskeletal structure. J Cell Biol 1985; 100: 1209-1218.
21. Liu G, Grant WM, Persky D, Latham VM Jr, Singer RH, Condeelis J. Interactions of elongation factor 1alpha with F-actin and beta-actin mRNA: implications for anchoring mRNA in cell protrusions. Mol Biol Cell 2002; 13: 579-592.
22. Hollenberg AN, Monden T, Madura JP, Lee K, Wondisford FE. Function of nuclear co-repressor protein on thyroid hormone response elements is regulated by the receptor A/B domain. J Biol Chem 1996; 271: 28516-28520.
23. Levada-Pires AC, Fonseca CE, Hatanaka E, Alba-Loureiro T, D'Angelo A, Velhote FB, et al. The effect of an adventure race on lymphocyte and neutrophil death. Eur J Appl Physiol 2010; 109: 447-453.
24. Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987; 162: 156-159.
25. Salles FJ, Richards WG, Strickland S. Assaying the polyadenylation state of mRNAs. Methods 1999; 17: 38-45.
26. Serrano-Nascimento C, Calil-Silveira J, Nunes MT. Posttranscriptional regulation of sodium-iodide symporter mRNA expression in the rat thyroid gland by acute iodide administration. Am J Physiol Cell Physiol 2010; 298: C893-C899.
27. Posern G, Sotiropoulos A, Treisman R. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol Biol Cell 2002; 13: 4167-4178.
28. Haller K, Rambaldi I, Daniels E, Featherstone M. Subcellular localization of multiple PREP2 isoforms is regulated by actin, tubulin, and nuclear export. J Biol Chem 2004; 279: 49384-49394.
29. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976; 72: 248-254.
30. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227: 680-685.
31. Carneiro I, Castro-Piedras I, Munoz A, Labandeira-Garcia JL, Devesa J, Arce VM. Hypothyroidism is associated with increased myostatin expression in rats. J Endocrinol Invest 2008; 31: 773-778.
32. Hovland R, Hesketh JE, Pryme IF. The compartmentalization of protein synthesis: importance of cytoskeleton and role in mRNA targeting. Int J Biochem Cell Biol 1996; 28: 1089-1105.
33. Mahon P, Partridge K, Beattie JH, Glover LA, Hesketh JE. The 3' untranslated region plays a role in the targeting of metallothionein-I mRNA to the perinuclear cytoplasm and cytoskeletal-bound polysomes. Biochim Biophys Acta 1997; 1358: 153-162.
34. Shannon JM, Pan T, Edeen KE, Nielsen LD. Influence of the cytoskeleton on surfactant protein gene expression in cultured rat alveolar type II cells. Am J Physiol 1998; 274: L87-L96.
35. van den Berg A, Freitas J, Keles F, Snoek M, van Marle J, Jansen HM, et al. Cytoskeletal architecture differentially controls post-transcriptional processing of IL-6 and IL-8 mRNA in airway epithelial-like cells. Exp Cell Res 2006; 312: 1496-1506.
36. Li Z, Pandit S, Deutscher MP. Polyadenylation of stable RNA precursors in vivo. Proc Natl Acad Sci U S A 1998; 95: 12158-12162.
The authors thank Leonice Lourenço Poyares for excellent technical assistance, Dr. Adriana C. Levada-Pires and Wilson Kowabara for the help with the FACSCalibur flow cytometer and Dr. Renato Salgado for revising the English text. Research supported by FAPESP. F. Goulart-Silva and C. Serrano-Nascimento are recipients of FAPESP (#08/56446-9 and #09/50175-6, respectively) fellowships. M.T. Nunes is the recipient of a CNPq fellowship.
 Address for correspondence: M.T. Nunes, Departamento de Fisiologia e Biofísica, Instituto de Ciências Biomédicas, USP, 05508-900, São Paulo, SP, Brasil. Fax: +55-11-3091-7285. E-mail: mtnunes@icb.usp.br
Address for correspondence: M.T. Nunes, Departamento de Fisiologia e Biofísica, Instituto de Ciências Biomédicas, USP, 05508-900, São Paulo, SP, Brasil. Fax: +55-11-3091-7285. E-mail: mtnunes@icb.usp.br
Received April 14, 2011. Accepted August 31, 2011. Available online September 16, 2011. Published October 10, 2011.
The Brazilian Journal of Medical and Biological Research is partially financed by
All the contents of this journal, except where otherwise noted, is licensed under a Creative Commons Attribution License
- 1. Bergh JJ, Lin HY, Lansing L, Mohamed SN, Davis FB, Mousa S, et al. Integrin alphaVbeta3 contains a cell surface receptor site for thyroid hormone that is linked to activation of mitogen-activated protein kinase and induction of angiogenesis. Endocrinology 2005; 146: 2864-2871.
- 2. Cheng SY, Gong QH, Parkison C, Robinson EA, Appella E, Merlino GT, et al. The nucleotide sequence of a human cellular thyroid hormone binding protein present in endoplasmic reticulum. J Biol Chem 1987; 262: 11221-11227.
- 3. Psarra AM, Solakidi S, Sekeris CE. The mitochondrion as a primary site of action of steroid and thyroid hormones: presence and action of steroid and thyroid hormone receptors in mitochondria of animal cells. Mol Cell Endocrinol 2006; 246: 21-33.
- 4. Yen PM. Physiological and molecular basis of thyroid hormone action. Physiol Rev 2001; 81: 1097-1142.
- 5. Bhargava M, Lei J, Mariash CN, Ingbar DH. Thyroid hormone rapidly stimulates alveolar Na,K-ATPase by activation of phosphatidylinositol 3-kinase. Curr Opin Endocrinol Diabetes Obes 2007; 14: 416-420.
- 6. Kim B. Thyroid hormone as a determinant of energy expenditure and the basal metabolic rate. Thyroid 2008; 18: 141-144.
- 7. Kaminsky P, Klein M, Duc M. [Control of muscular bioenergetics by the thyroid hormones]. Presse Med 1993; 22: 774-778.
- 8. Lukens FD. Insulin and protein metabolism. Diabetes 1964; 13: 451-461.
- 9. Moller N, Jorgensen JO. Effects of growth hormone on glucose, lipid, and protein metabolism in human subjects. Endocr Rev 2009; 30: 152-177.
- 10. Foufelle F, Hegarty B, Bobard A, Hainault I, Bossard P, Ferre P. [A new role for insulin in the control of energetic metabolism in liver]. Med Sci 2005; 21: 569-571.
- 11. Vijayakumar A, Novosyadlyy R, Wu Y, Yakar S, LeRoith D. Biological effects of growth hormone on carbohydrate and lipid metabolism. Growth Horm IGF Res 2010; 20: 1-7.
- 12. Misiti S, Anastasi E, Sciacchitano S, Verga FC, Panacchia L, Bucci B, et al. 3,5,3'-Triiodo-L-thyronine enhances the differentiation of a human pancreatic duct cell line (hPANC-1) towards a beta-cell-like phenotype. J Cell Physiol 2005; 204: 286-296.
- 13. Fernandez-Mejia C, Davidson MB. Regulation of glucokinase and proinsulin gene expression and insulin secretion in RIN-m5F cells by dexamethasone, retinoic acid, and thyroid hormone. Endocrinology 1992; 130: 1660-1668.
- 14. Fukuchi M, Shimabukuro M, Shimajiri Y, Oshiro Y, Higa M, Akamine H, et al. Evidence for a deficient pancreatic beta-cell response in a rat model of hyperthyroidism. Life Sci 2002; 71: 1059-1070.
- 15. Yildiz BO, Haznedaroglu IC. Rethinking leptin and insulin action: therapeutic opportunities for diabetes. Int J Biochem Cell Biol 2006; 38: 820-830.
- 16. Holst JJ. The physiology of glucagon-like peptide 1. Physiol Rev 2007; 87: 1409-1439.
- 17. Strowski MZ, Parmar RM, Blake AD, Schaeffer JM. Somatostatin inhibits insulin and glucagon secretion via two receptor subtypes: an in vitro study of pancreatic islets from somatostatin receptor 2 knockout mice. Endocrinology 2000; 141: 111-117.
- 18. Silva FG, Giannocco G, Luchessi AD, Curi R, Nunes MT. T3 acutely increases GH mRNA translation rate and GH secretion in hypothyroid rats. Mol Cell Endocrinol 2010; 317: 1-7.
- 19. Goulart-Silva F, de Souza PB, Nunes MT. T3 rapidly modulates TSHbeta mRNA stability and translational rate in the pituitary of hypothyroid rats. Mol Cell Endocrinol 2011; 332: 277-282.
- 20. Bonneau AM, Darveau A, Sonenberg N. Effect of viral infection on host protein synthesis and mRNA association with the cytoplasmic cytoskeletal structure. J Cell Biol 1985; 100: 1209-1218.
- 21. Liu G, Grant WM, Persky D, Latham VM Jr, Singer RH, Condeelis J. Interactions of elongation factor 1alpha with F-actin and beta-actin mRNA: implications for anchoring mRNA in cell protrusions. Mol Biol Cell 2002; 13: 579-592.
- 22. Hollenberg AN, Monden T, Madura JP, Lee K, Wondisford FE. Function of nuclear co-repressor protein on thyroid hormone response elements is regulated by the receptor A/B domain. J Biol Chem 1996; 271: 28516-28520.
- 23. Levada-Pires AC, Fonseca CE, Hatanaka E, Alba-Loureiro T, D'Angelo A, Velhote FB, et al. The effect of an adventure race on lymphocyte and neutrophil death. Eur J Appl Physiol 2010; 109: 447-453.
- 24. Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987; 162: 156-159.
- 25. Salles FJ, Richards WG, Strickland S. Assaying the polyadenylation state of mRNAs. Methods 1999; 17: 38-45.
- 26. Serrano-Nascimento C, Calil-Silveira J, Nunes MT. Posttranscriptional regulation of sodium-iodide symporter mRNA expression in the rat thyroid gland by acute iodide administration. Am J Physiol Cell Physiol 2010; 298: C893-C899.
- 27. Posern G, Sotiropoulos A, Treisman R. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol Biol Cell 2002; 13: 4167-4178.
- 28. Haller K, Rambaldi I, Daniels E, Featherstone M. Subcellular localization of multiple PREP2 isoforms is regulated by actin, tubulin, and nuclear export. J Biol Chem 2004; 279: 49384-49394.
- 29. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976; 72: 248-254.
- 30. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227: 680-685.
- 31. Carneiro I, Castro-Piedras I, Munoz A, Labandeira-Garcia JL, Devesa J, Arce VM. Hypothyroidism is associated with increased myostatin expression in rats. J Endocrinol Invest 2008; 31: 773-778.
- 32. Hovland R, Hesketh JE, Pryme IF. The compartmentalization of protein synthesis: importance of cytoskeleton and role in mRNA targeting. Int J Biochem Cell Biol 1996; 28: 1089-1105.
- 33. Mahon P, Partridge K, Beattie JH, Glover LA, Hesketh JE. The 3' untranslated region plays a role in the targeting of metallothionein-I mRNA to the perinuclear cytoplasm and cytoskeletal-bound polysomes. Biochim Biophys Acta 1997; 1358: 153-162.
- 34. Shannon JM, Pan T, Edeen KE, Nielsen LD. Influence of the cytoskeleton on surfactant protein gene expression in cultured rat alveolar type II cells. Am J Physiol 1998; 274: L87-L96.
- 35. van den Berg A, Freitas J, Keles F, Snoek M, van Marle J, Jansen HM, et al. Cytoskeletal architecture differentially controls post-transcriptional processing of IL-6 and IL-8 mRNA in airway epithelial-like cells. Exp Cell Res 2006; 312: 1496-1506.
- 36. Li Z, Pandit S, Deutscher MP. Polyadenylation of stable RNA precursors in vivo Proc Natl Acad Sci U S A 1998; 95: 12158-12162.
Correspondence and Footnotes
Publication Dates
-
Publication in this collection
11 Oct 2011 -
Date of issue
Oct 2011
History
-
Accepted
31 Aug 2011 -
Received
14 Apr 2011